Abstract
The FDA-approved prophylactic antimalarial drug atovaquone (ATO) recently was repurposed as an antitumor drug. Studies show that ATO exerts a profound antiproliferative effect in several cancer cells, including breast, ovarian, and glioma. Analogous to the mechanism of action proposed in parasites, ATO inhibits mitochondrial complex III and cell respiration. To enhance the chemotherapeutic efficacy and oxidative phosphorylation inhibition, we developed a mitochondria-targeted triphenylphosphonium-conjugated ATO with varying alkyl side chains (Mito4-ATO, Mito10-ATO, Mito12-ATO, and Mito16-ATO). Results show, for the first time, that triphenylphosphonium-conjugated ATO potently enhanced the antiproliferative effect of ATO in cancer cells and, depending upon the alkyl chain length, the molecular target of inhibition changes from mitochondrial complex III to complex I. Mito4-ATO and Mito10-ATO inhibit both pyruvate/malate-dependent complex I and duroquinol-dependent complex III-induced oxygen consumption whereas Mito12-ATO and Mito16-ATO inhibit only complex I-induced oxygen consumption. Mitochondrial target shifting may have immunoregulatory implications.
Subject terms: Breast cancer, Lung cancer, Pancreatic cancer, NMR spectroscopy, Biological techniques, Biophysics, Drug discovery, Molecular biology, Molecular medicine
Introduction
Atovaquone (ATO), a hydroxy-1,4-naphthoquinone analog of ubiquinone (Q), also known as coenzyme Q10 (Fig. 1A), is an FDA-approved antimicrobial drug used to treat pneumocystis pneumonia and to prevent and treat malaria caused by the parasites Pnemocystis jirovecii and Plasmodium falciparum and toxoplasmosis infections in immune-compromised HIV patients1,2. ATO exerts antiviral effects, inhibiting arboviruses3. ATO is the first clinically approved drug that targets Plasmodium cytochrome bc1 complex in mitochondria4. Also, ATO acts as a competitive inhibitor of mitochondrial complex III by displacing ubiquinol at the active site of the cytochrome bc1 complex, inhibiting mitochondrial respiration and mitochondrial membrane potential in parasites and killing them5.
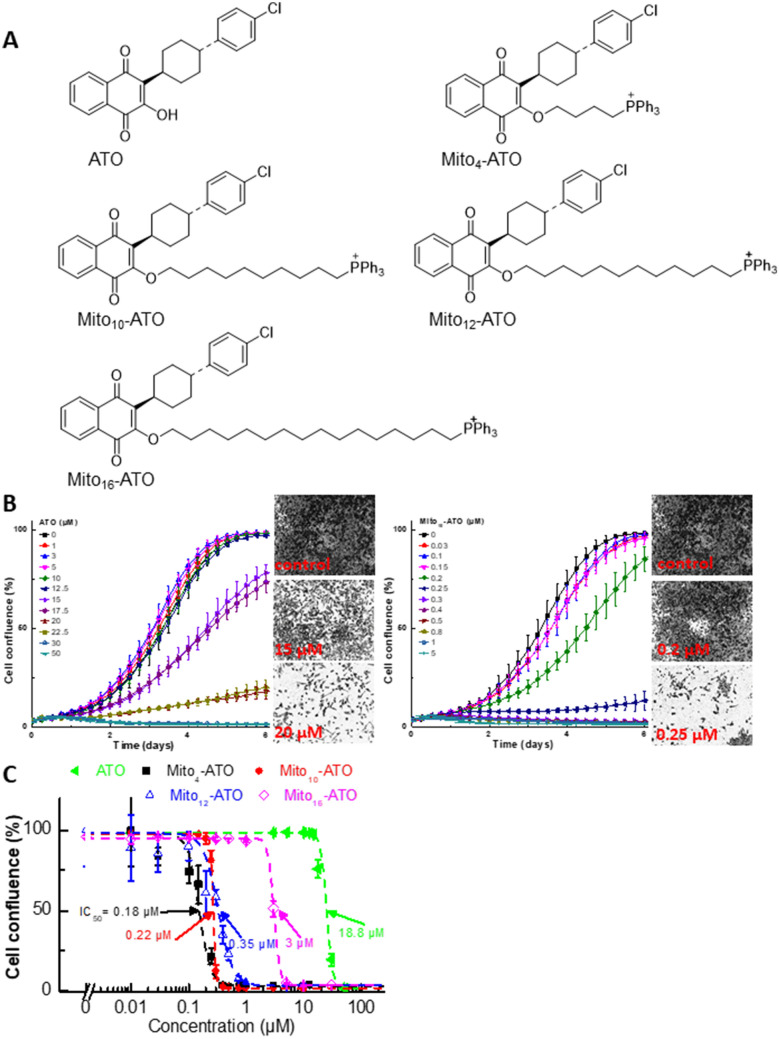
Figure 1.
Effects of ATO, Mito-ATO analogs, and related analogs on proliferation of MiaPaCa-2 cells. (A) Chemical structures of ATO and Mito-ATO analogs. (B) Effect of ATO on the proliferation of human pancreatic cancer cells was compared with that of Mito10-ATO in the IncuCyte Live-Cell Imager. MiaPaCa-2 cells were treated with ATO and Mito10-ATO. Cell proliferation was monitored in real-time with the continuous presence of indicated treatments until the end of each experiment. Created using NIH public domain image processing program, ImageJ38. (C) Cell confluence (as control groups reach 98% confluency) is plotted against concentrations of ATO and Mito-ATO analogs. Dashed lines represent the fitting curves used to determine their IC50 values as indicated.
Recently, ATO was repurposed to target mitochondrial complex III in breast cancer cells, and results show that ATO inhibits proliferation of breast cancer stem-like cells6. The trans form of ATO is significantly more potent than the cis form. A similar mechanism of action (mitochondrial complex III inhibition) was proposed in breast cancer cells6, although the affinity of ATO to mitochondrial cytochrome bc1 complex in mammalian cells is much lower than in parasites7. Targeting of ATO to mitochondrial complex III in ovarian cancer cells as a potential antitumor therapeutic strategy was proposed8. More recent studies show that ATO or ATO and proguanil (i.e., Malarone) exhibit antitumor activity both in animal models and in patients with acute myelogenous leukaemia and acute lymphocytic leukaemia9. Inhibition of tumour growth by ATO was attributed to inhibition of phosphorylation of signal transducer and activator of transcription 3 (STAT3)10. ATO has been shown to inhibit glioblastoma cell proliferation, and inhibition of STAT3 by ATO as a viable therapy for glioblastoma multiforme was proposed10. However, the ATO concentration in the brain was suggested to be too low to be chemotherapeutically effective.
We and others have previously shown that compared to their untargeted analogs, the triphenylphosphonium (TPP+)-conjugated derivatives are typically much more potent (> 50–100 times) in inhibiting tumour cell proliferation11–14. TPP+-conjugated mitochondria-targeted compounds are selectively taken into cancer cells at much higher levels due to an increased negative mitochondrial membrane potential11,12. The antiproliferative potency of TPP+-modified metformin, attributed to mitochondrial complex I inhibition, increased with increasing aliphatic side chain length11,12. Both metformin and mito-metformin analogs inhibit mitochondrial complex I-mediated respiration. Little or no information exists on TPP + -modified complex III inhibitors. Thus, we modified the structure of ATO, an established complex III inhibitor, and developed TPP+-conjugated ATO (Fig. 1A) and investigated their antiproliferative and oxidative phosphorylation (OXPHOS) inhibitory effects in cancer cells.
The potent inhibition of mitochondrial complex III may have implications in the maintenance of the immunosuppressive function of regulatory T (Treg) cells15. Although several relatively nontoxic mitochondrial complex I inhibitors16 exist (other than rotenone [Rot], which is toxic), antimycin A is one of the few complex III inhibitors presently available. Developing potent mitochondrial complex III inhibitors is timely because of their ability to suppress Treg cells and enhance the levels of effector T (Teff) cells15.
In the present study, we show that mitochondria-targeted ATO (Mito-ATO) analogs (Fig. 1) are significantly more potent than ATO in inhibiting pancreatic cancer cell proliferation. Our results also show, for the first time, that conjugating ATO to TPP+ and increasing the aliphatic side chain length switches the molecular target in mitochondria from complex III/complex I to complex I for Mito-ATO analogs. As a result, Mito12-ATO and Mito16-ATO block mitochondrial respiration in pancreatic cancer cells by inhibiting only complex I and not complex III, whereas Mito4-ATO and Mito10-ATO inhibit oxygen consumption induced by both mitochondrial complex I and complex III. Potential implications of enhanced complex III inhibition induced by Mito4-ATO and Mito12-ATO in cancer immunosuppression are discussed.
Results
Mito-ATO analogs are more potent than ATO in inhibiting MiaPaCa-2 pancreatic cancer cell proliferation
Cell proliferation was monitored continuously in real time using an IncuCyte image analyser11–13 and both ATO and Mito-ATO analogs (Fig. 1A). Figure 1B shows the dose-dependent antiproliferative effects of ATO and Mito10-ATO. Mito4-ATO, Mito12-ATO, and Mito16-ATO also dose-dependently inhibited proliferation of MiaPaCa-2 pancreatic cancer cells. Mito-ATO analogs are more potent than ATO at inhibiting the proliferation of MiaPaCa-2 cells. Figure 1C shows the cell confluence (indicated by a dotted line) as a function of Mito-ATO concentration, and the half maximal inhibitory concentration (IC50) values of Mito4-ATO, Mito10-ATO, Mito12-ATO, and Mito16-ATO are 0.18 µM, 0.22 µM, 0.35 µM, and 3 µM, respectively. As compared with Mito-ATO analogs, ATO inhibited cell proliferation at much higher concentrations (IC50 = 18 µM) (Fig. 1C). These results suggest that attaching an aliphatic chain containing a TPP+ group to ATO greatly increases the antiproliferative potency. In control experiments, we used compounds (butyl-ATO and decyl-ATO) with an alkyl carbon–carbon side chain length similar to those of Mito4-ATO and Mito10-ATO but lacking the TPP+. As shown in Figure S3, both butyl-ATO and decyl-ATO devoid of TPP+ were much less effective than their corresponding Mito-ATO analogs.
Inhibitory effects of Mito-ATO analogs on mitochondrial complex activities in MiaPaCa-2 cells
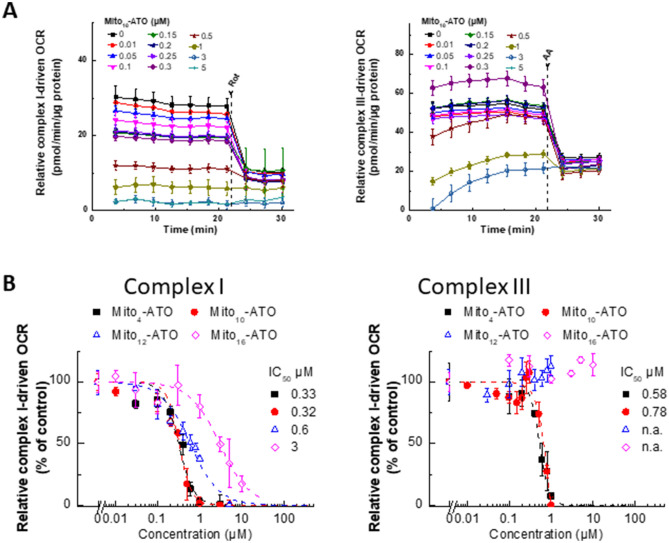
The mitochondrial complex activities were assessed by measuring the oxygen consumption rate (OCR) using the Seahorse technique6,11–14. MiaPaCa-2 cells were treated separately with Mito-ATO analogs at different concentrations for 24 h and OCR was measured. As shown in Fig. 2A, Mito10-ATO effectively inhibits complex III-induced oxygen consumption and, more importantly, Mito10-ATO caused a significantly greater inhibition of mitochondrial complex I-driven oxygen consumption. The IC50 value for Mito10-ATO to inhibit oxygen consumption by complex I is 0.32 µM, and the IC50 value for Mito10-ATO to inhibit oxygen consumption by complex III is 0.78 µM (Fig. 2B). Mito4-ATO and Mito10-ATO potently inhibit both mitochondrial complex I- and complex III-induced oxygen consumption. However, Mito12-ATO and Mito16-ATO did not inhibit complex III-induced oxygen consumption; they inhibited only the complex I-induced oxygen consumption (Fig. 2B). These results show that there is a shift in mitochondrial targeting of Mito-ATO analogs that is dependent on the alkyl side chain length attached to TPP+.
Figure 2.
Effects of Mito-ATO analogs on the oxygen consumption induced by mitochondrial complexes I and III. (A) Dose-dependent effect of Mito10-ATO on complex I- and complex III-dependent oxygen consumption was measured in MiaPaCa-2 cells. MiaPaCa-2 cells were treated with Mito-ATO for 24 h. Relative complex I (1.5 mM malate, 10 mM pyruvate, 10 mM ADP)-driven OCR (left) and relative complex III (0.5 mM duroquinol, 10 mM ADP)-driven OCR (right) were monitored by XF-96 analyser. Either Rot (complex I inhibitor) or antimycin A (AA, complex III inhibitor) was acutely added and OCR assayed immediately. (B) The mitochondrial complex I (left)- and III (right)-driven OCR (calculated as Rot or AA inhibitable OCR) are plotted against the concentration of ATO and Mito-ATO analogs. Dashed lines represent the fitting curves used for determination of the IC50 values. (n.a., not applicable).
Relative uptake of Mito10-ATO and ATO into MiaPaCa-2 and A549 cells
Next, we used the liquid chromatography with tandem mass spectrometry (LC–MS/MS) technique to investigate the relative uptake of Mito10-ATO and ATO in MiaPaCa-2 and A549 cells (Fig. S4). Cells were treated with Mito10-ATO (0.1 µM and 1 µM) or ATO (1–10 µM) for 1, 2, 4, and 24 h. As shown in Figure S4, there was an increase in cellular uptake of Mito10-ATO that accumulated inside the cells with time. Under the same treatment period with even with higher concentrations (up to 10 µM), ATO uptake was considerably lower and did not accumulate in cancer cells over a 24 h period.
Inhibitory effects of Mito10-ATO on proliferation of breast and lung cancer cells
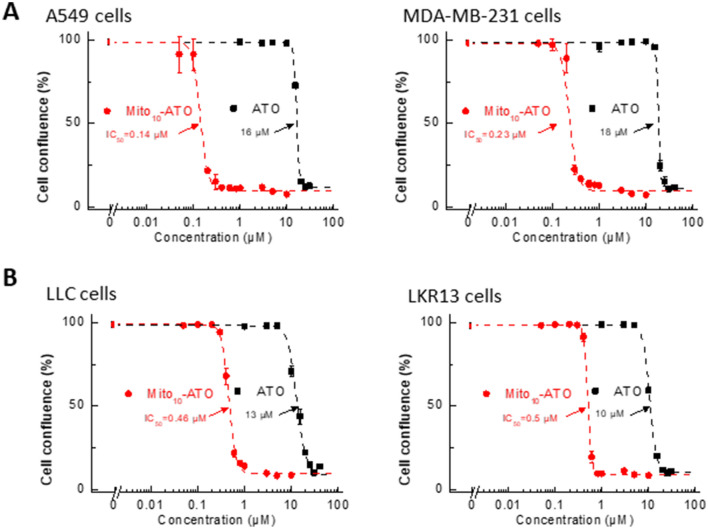
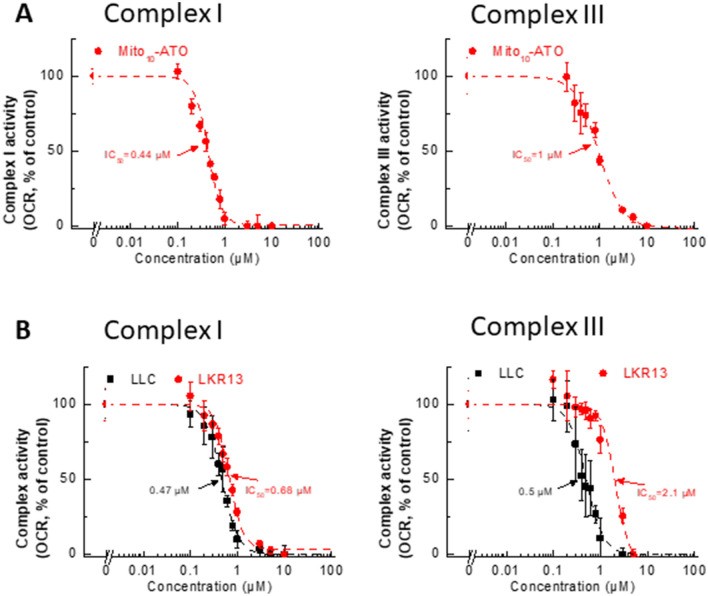
Previous reports provide evidence for enhanced uptake of several mitochondria-targeted cationic agents into cancer cells as compared with non-transformed control cells11–14. We examined the effect of ATO and Mito10-ATO on A549 human lung cancer cells and MDA-MB-231 human breast cancer cells (Fig. 3A) and on LLC and LKR13 mouse lung cancer cells (Fig. 3B). Mito10-ATO was effective at halting cell proliferation in A549 and MDA-MB-231 human cancer cells and in LLC and LKR13 mouse lung cancer cells. As shown in Fig. 3, Mito10-ATO also was more potent than ATO in inhibiting cancer cell proliferation. Mito10-ATO inhibited both mitochondrial complex I- and III-induced oxygen consumption in these cells (Fig. 4). These results demonstrate that the enhanced antiproliferative potency of Mito-ATO analogs is not restricted to a single cancer cell type and is broadly applicable to several cancer cells.
Figure 3.
Effects of ATO and Mito-ATO on proliferation in human cancer cells and mouse cancer cells. A549 human lung and MDA-MB-231 human breast cancer cells (A) and LLC and LKR13 mouse lung cancer cells (B) were treated with ATO and Mito10-ATO. Cell proliferation was monitored in real-time with the continuous presence of indicated treatments until the end of each experiment. The cell confluence (as control groups reach 98% confluency) is plotted against concentration. Dashed lines represent the fitting curves used to determine the IC50 values as indicated.
Figure 4.
Effects of Mito10-ATO on the oxygen consumption induced by mitochondrial complexes I and III in human cancer cells and mouse cancer cells. A549 human lung cancer cells (A), and LLC and LKR13 mouse lung cancer cells (B) were treated with Mito10-ATO. The mitochondrial complex I (left) and complex III (right) -driven OCR (calculated as Rot or AA inhibitable OCR) are plotted against concentration of Mito10-ATO. Dashed lines represent the fitting curves used for determination of the IC50 values.
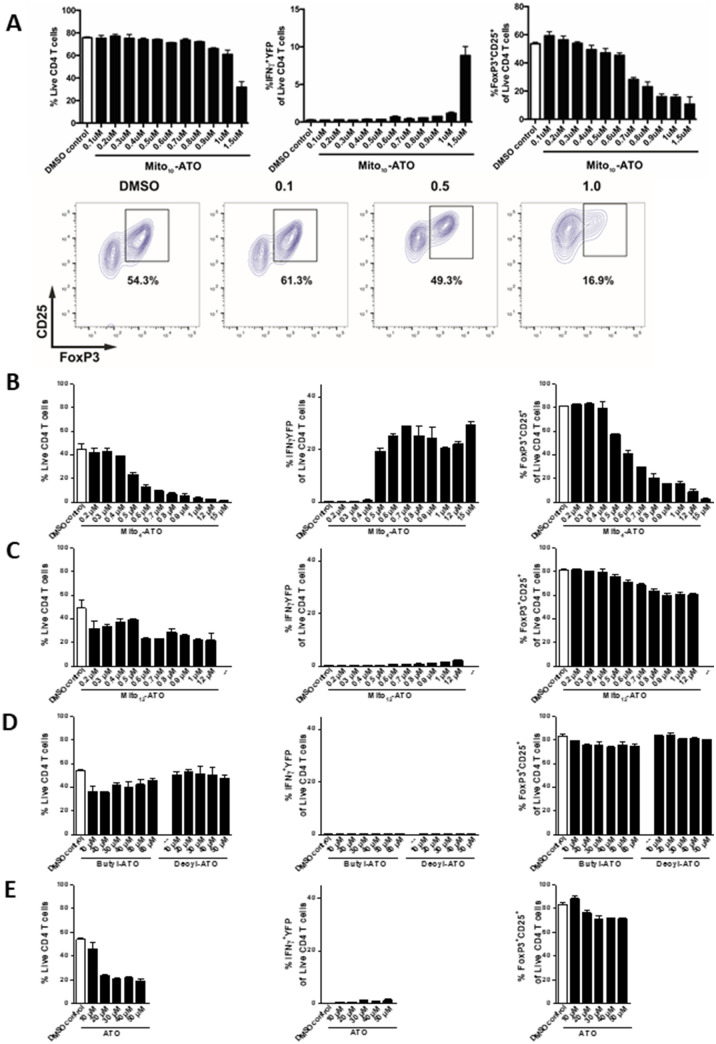
Immunomodulatory effects of Mito4-ATO, Mito10-ATO, and Mito12-ATO
Recent reports indicate that selective targeting and inhibiting of mitochondrial complex III mitigate and reverse immunosuppression by Treg cells, promoting the function of Teff cells15,17. To investigate the effects of Mito4-ATO, Mito10-ATO, and Mito12-ATO on Teff versus Treg cells, activated CD4+ T cells were isolated from SMARTA triple reporter mice, activated, and cultured in vitro with TGFβ (5 ng/mL) and IL-2 (100 ug/mL), as described in the Materials and Methods section. The CD4+ T cells were treated with ATO and Mito-ATO analogs at varying concentrations (Fig. 5). After six days, cells were stained to assess viability, phenotype, and function using flow cytometry. Results demonstrate that Mito4-ATO (Fig. 5B) and Mito10-ATO (Fig. 5A) inhibited Foxp3+ Treg differentiation and/or survival and promoted Teff cell IFNγ production in a dose-dependent manner. In contrast, Mito12-ATO did not appreciably inhibit Treg differentiation (Fig. 5C). Mito4-ATO and Mito10-ATO potently inhibited mitochondrial complex I- and complex III-driven oxygen consumption. Mito12-ATO strongly inhibited oxygen consumption by complex I- but not complex III-driven oxygen consumption. Thus, it is plausible that inhibition of Treg and stimulation of Teff response by Mito4-ATO and Mito10-ATO are mediated by their increased potency to target mitochondrial complex III. Furthermore, ATO, butyl-ATO, and decyl-ATO (up to 60 µM) did not inhibit Treg differentiation and/or survival (Fig. 5D, E).
Figure 5.
In vitro differentiation of CD4+ T regulatory cells under varying concentrations of Mito-ATOs and controls. After six days of culture, cells were stained for flow cytometry analysis. (left) Live/dead staining to assess the percentage of live CD4 + T cells within the lymphocyte gate. (middle) Teff cell function is shown as the frequency of IFNγ-YFP positive cells within the live CD4+ T cells. (right) The percentage of Treg cells is shown as the frequency of FoxP3+CD25+ cells within the live CD4+ T cells. (A, bottom) Representative plot using FlowJo Software, effects of Mito10-ATO on Treg (FoxP3+CD25+) cells within the live CD4+ T cell gate (contour plots depict gating corresponding to panel A, upper right)39. Mito4-ATO (B) effectively suppressed Treg while Mito12-ATO (C), Butyl-ATO (D), Decyl-ATO (D), and ATO (E) did not suppress Treg cells.
Discussion
The relative hydrophobicity of ATO, Mito-ATO, and alkyl-ATO
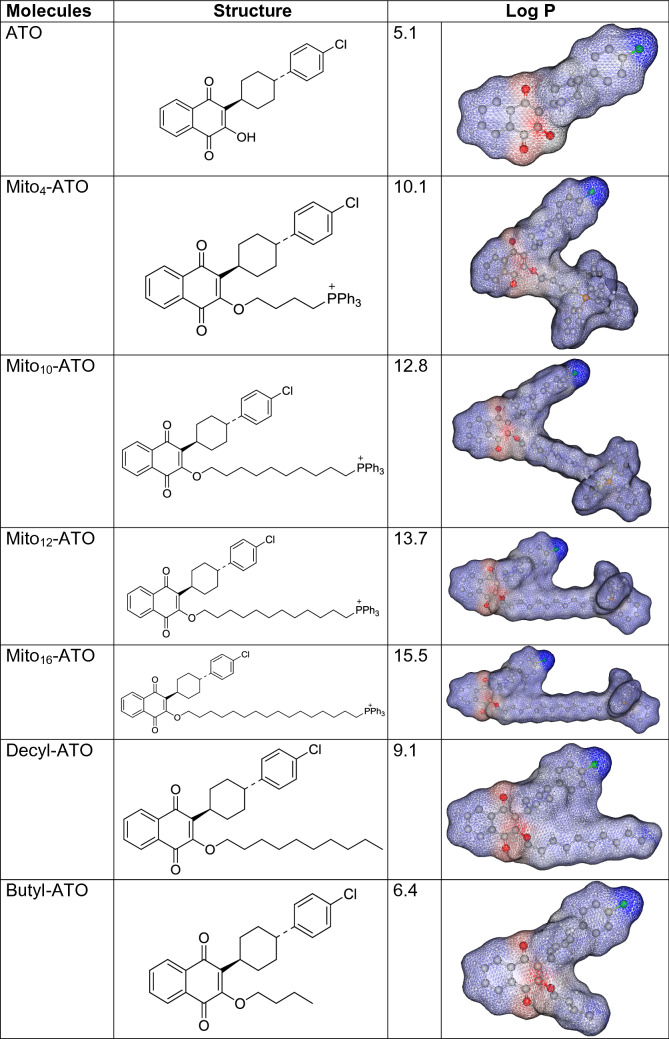
The lipophilicity of ATO was attributed to its stabilization in the hydrophobic pocket of the cytochrome bc1 complex5. To assess the relative hydrophobicity of ATO and Mito-ATO analogs, we calculated the octanol/water partition coefficients (log P) using a QSAR analysis and rational drug design as a measure of molecular hydrophobicity (Table 1). This method also uses a consensus model built using the ChemAxon software (San Diego, CA)18,19. Table 1 lists the log P values along with the calculated regions of the relative hydrophilic and hydrophobic regions. As shown in Table 1, there was a significant increase in the hydrophobicity of Mito-ATO (as compared with ATO) with the increasing alkyl side chain length (from C-4 to C-16).
Table 1.
Calculated partition coefficients and relative hydrophobic regions in ATO and Mito-ATO analogs.
Inhibition of cytochrome bc1 by ATO: Stabilizing molecular interactions at the active site
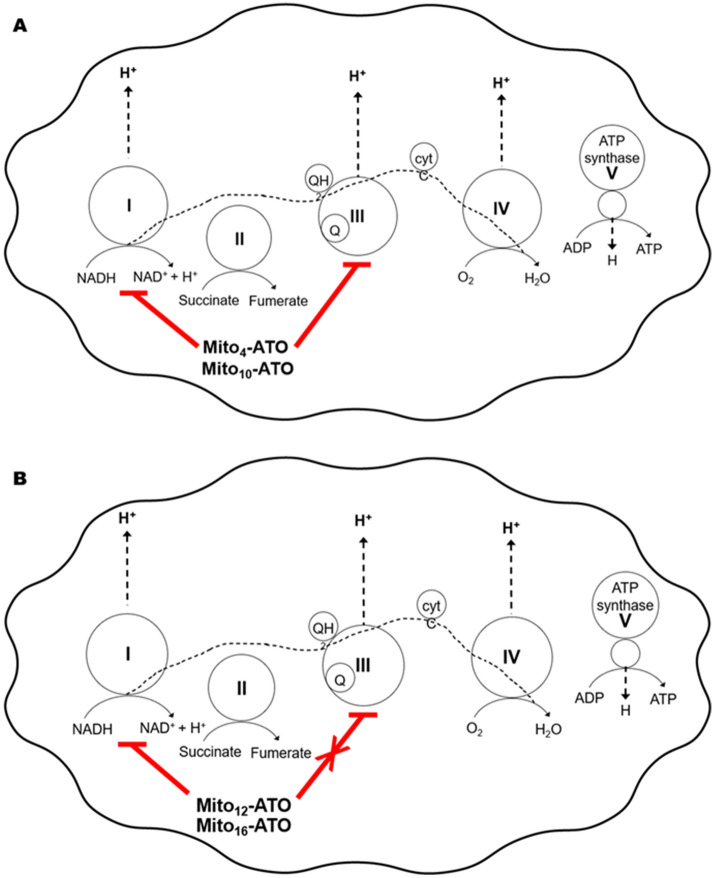
Previous studies showed that ATO binds to the mitochondrial cytochrome bc1 complex (ubiquinol cytochrome c oxidoreductase or complex III) and inhibits its activity5,20. We report here that structural modification of ATO by attachment of TPP+ to ATO (Mito-ATOs) greatly inhibits tumour cell proliferation. Both Mito4-ATO and Mito10-ATO potently inhibit oxygen consumption by complex I and complex III (Fig. 6A). Surprisingly, Mito12-ATO (with a 12-carbon side chain) and Mito16-ATO (with a 16-carbon side chain) only inhibited complex I- but not complex III-driven oxygen consumption (Fig. 6B). Conceivably, the lack of effect of Mito12-ATO and Mito16-ATO on oxygen consumption by complex III suggests that Mito12-ATO and Mito16-ATO do not target the Qo site of the cytochrome bc1 complex.
Figure 6.
Mitochondrial targeting of Mito-ATOs with varying side chain lengths. This figure illustrates (A) the dual mitochondrial targeting (complex I and complex III) of Mito4-ATO and Mito10-ATO and (B) a shift in mitochondrial targeting from complex III to complex I for Mito12-ATO and Mito16-ATO in MiaPaCa-2 cells.
ATO is structurally similar to Q and acts as a competitive inhibitor of Q. Modelling and energy minimization studies show that ATO docks into the Qo active site stabilized by hydrophobic and hydrogen bonding interactions with the Rieske protein and the cytochromes5,21. Q is reduced to ubiquinol (QH2) by complexes I and II and oxidized to Q at the catalytic site of complex III. Based on published reports on the molecular basis of the antimalarial action of ATO5, a similar mechanism is proposed for ATO inhibition of respiration of cancer cells. ATO acts as a competitive Qo site-specific inhibitor of cytochrome bc1 complex oriented between the [2Fe–2S] cluster of the Rieske protein. ATO is stabilized in this pocket through the polarized hydrogen bonding interaction between the oxygen atom (O) of the ionized form and the protonated nitrogen atom of the His181 side chain of the Rieske protein.
The evidence for the binding of ATO to the Rieske iron-sulphur centre came from electron paramagnetic resonance (EPR) spectroscopy and circular dichroism spectroscopy studies of the purified cytochrome bc1 complex from the yeast Saccharomyces cerevisiae4. The addition of ATO shifted the ascorbate-reduced Rieske centre signals (at gz and gx to a higher field and the gy component to a lower field). The extent of the magnetic field shifts in the EPR spectra of the reduced Rieske Fe-S centre indicates a change in the electronic environment of the 2Fe-2S cluster due to ATO binding4. The low-temperature EPR of the yeast system may be suitable for investigating the effect of alkyl side chain length in Mito-ATO (e.g., Mito4-ATO, Mito10-ATO, Mito12-ATO, or Mito16-ATO) and the Rieske iron-sulphur cluster.
It is plausible that, depending on the length of the aliphatic substituent attached to the TPP+ group in Mito-ATO, some Mito-ATO analogs (Mito4-ATO or Mito10-ATO) may be stabilized at the cytochrome bc1 pocket by several hydrophobic/aromatic interactions between the ATO moiety and the amino acid residues within the binding site. Mito-ATO is much more hydrophobic than ATO. It accumulates into cancer cell mitochondria more effectively than ATO and inhibits mitochondrial respiration and cancer cell proliferation more potently than ATO. Results using Mito12-ATO indicate that it does not inhibit complex III-dependent oxygen consumption. However, Mito12-ATO potently inhibits complex I-driven oxygen consumption (Fig. 6). This, combined with the finding that Mito12-ATO does not inhibit Treg cells, implies that the alkyl side chain length in Mito-ATO plays an important role in mitochondrial targeting and immunoregulatory effects22.
Cancer immunosuppression and Mito-ATO
The number of Treg cells is reportedly increased in the microenvironments of most cancers (e.g., pancreatic cancer, lung cancer)23,24. There is negative correlation between Treg levels and survival in cancer patients25. Treg cells suppress antitumor immunity, thereby hampering immunotherapy26. Drug therapy targeting Treg cells is emerging as a promising antitumor approach27,28. Emerging research shows that mitochondrial respiratory chain activity, particularly mitochondrial complex III activity, is crucial for preserving the antitumor function of Treg cells29. Alternatively, Treg cells devoid of complex III had decreased immunoregulatory function15. It is conceivable that Mito-ATO analogs with the appropriate substituents and aliphatic side chain lengths will be able to inhibit Treg cell respiration and activate cancer immunotherapy.
Clearly, the discovery that increasing alkyl chain length in Mito-ATO, a new class of small-molecule OXPHOS inhibitors, changes the mitochondrial respiratory complex target is significant in corroborating the role of complex III-dependent metabolic alterations (i.e., accumulation of oncometabolite 2-hydroxyglutarate [2-HG] and succinate) in repressing alpha-ketoglutarate-dependent demethylases and DNA hypermethylation15. Previously, it was shown that loss or inhibition of mitochondrial complex III in cancer cells results in increased levels of 2-HG and succinate15,17. Antimycin A increased the levels of succinate and 2-HG in Treg cells15. Our initial attempts to measure mitochondrial complex activities, using the Seahorse technique, in intact and permeabilized T cells were not successful. Several experimental conditions (e.g., cell concentration, permeabilizing conditions) need to be optimized before reliable oxygen consumption rate measurements can be made. To further corroborate the role of mitochondrial complex III, it would be of interest to compare the effects of ATO, Mito4-ATO, Mito10-ATO, Mito12-ATO, and Mito16-ATO on inducing the transcriptional programs responsible for metabolic alterations in cancer cells and immune cells15,17.
In this study, we showed that TPP+-conjugated ATO analogs (Mito4-ATO, Mito10-ATO, Mito12-ATO, and Mito16-ATO) are considerably more potent than the parent drug, ATO, at inhibiting the proliferation of pancreatic and other cancer cell types. Mito4-ATO and Mito10-ATO but not Mito12-ATO suppress Treg function. The molecular targets of Mito-ATO analogs in the mitochondrial respiratory chain are different depending on the alkyl side chain length. Mito4-ATO and Mito10-ATO inhibit mitochondrial oxygen consumption by complex I and complex III, whereas Mito12-ATO and Mito16-ATO inhibit only the complex I-induced oxygen consumption. Interestingly, the alkyl side chain length in Mito-ATOs influences their mitochondrial targeting and inhibition of oxygen consumption.
Materials and methods
General
All chemicals and organic solvents were commercially available and were used as supplied. The reactions were monitored by thin layer chromatography using silica gel Merck 60F254. Crude materials were purified by flash chromatography on Merck silica gel 60 (0.040–0.063 mm). 31P NMR, 1H NMR and 13C NMR spectra were recorded at 400 and 75 MHz spectrometers, respectively. 1H NMR spectra were recorded using a Bruker DPX AVANCE 400 spectrometer equipped with a quattro nucleus probe. Chemical shifts (δ) are reported in ppm and J values in Hertz.
Syntheses and nuclear magnetic resonance (NMR) data of appropriate ATO controls lacking the TPP+ group are given in Figs. S1 and S2 in the Supplementary Materials.
Synthesis of Mito-ATO analogs
The mitochondria-targeted analogs of ATO (Miton-ATO) were prepared by reacting the appropriates bromoalkyl-triphenylphosphonium bromides with ATO in the presence of potassium carbonate in dimethylformamide (DMF) (Fig. S1). In addition, the untargeted ATO derivatives (ATO-Cn) were prepared by adapting the procedure to the corresponding alkyl bromides (Fig. S1).
Synthesis of Mito4-ATO
Mito4-ATO was prepared by reacting (4-bromobutyl)-triphenylphosphonium bromide with ATO in the presence of potassium carbonate in DMF (Fig. S1). Briefly, (4-bromobutyl)-triphenylphosphonium bromide (0.39 g, 0.81 mmol) was added to a mixture of ATO (0.3 g, 0.82 mmol) and potassium carbonate (0.15 g, 0.82 mmol) in DMF. The mixture was stirred at 70 °C for 9 h. CH2Cl2 was then added to the mixture followed by the addition of water (20 mL). The organic layer was washed twice with water and dried over Na2SO4. The solvent was removed under reduced pressure. Diethyl ether was added to the mixture to precipitate out the compound that was purified by flash chromatography (CH2Cl2/EtOH, 9:1), yielding the product, Mito4-ATO (0.47 g, 75% yield).
The HRMS calculated and found values are C44H41ClO3P+ [M+] 683.2476 and 683.2479. 31P NMR (400.13 MHz, CDCl3) δ 24.66. 1H NMR (400.13 MHz, CDCl3), δ 8.06–8.03 (1H, m), 7.95–7.93 (1H, m), 7.92–7.85 (6H, m), 7.79–7.72 (3H, m), 7.71–7.63 (8H, m), 7.25–7.21 (2H, m), 7.15–7.12 (2H, m), 4.26 (2H, t, J = 5.6), 4.11–4.02 (2H, m), 3.13–3.03 (1H, m), 2.58–2.48 (1H, m), 2.37–2.28 (2H, m), 2.10–1.98 (4H, m), 1.92–1.88 (2H, m), 1.65 (2H, dd, J = 12.7, 2.7), 1.54–1.39 (2H, m). 13C NMR (75 MHz, CDCl3) δ 185.4, 181.7, 157.7, 145.7, 140.0, 134.9, 134.8, 133.8, 133.77, 133.68, 133.2, 132.3, 131.4, 131.3, 130.5, 130.3, 128.3, 128.2, 118.8, 117.9, 72.4, 43.1, 35.5, 34.3, 30.12 (d, 17.6), 30.1, 30.0, 22.2 (d, J = 50.6), 19.1 (d, J = 3.7).
NMR spectra and related parameters are included in Fig. S2.
Synthesis of Mito10-ATO
Mito10-ATO was prepared by reacting (10-bromodecyl)-triphenylphosphonium bromide with ATO in the presence of potassium carbonate in DMF (Fig. S1). Briefly, (10-bromodecyl)-triphenylphosphonium bromide (1.1 g, 1.9 mmol) was added to a mixture of ATO (0.73 g, 1.9 mmol) and potassium carbonate (0.3 g, 2.1 mmol) in DMF (4 mL). The mixture was stirred at 70 °C for 9 h. Methylene chloride (CH2Cl2) was added to the mixture, and then water (20 mL) was added. The organic layer was washed twice with water and dried over anhydrous sodium sulphate. The excess solvent was removed under reduced pressure. Diethyl ether was subsequently added to the mixture to precipitate the compound. Purification by flash chromatography, using a mixture of methylene chloride and ethanol (9:1), yielded the desired compound, Mito10-ATO (1 g, 59% yield).
The high-resolution mass spectral (HRMS) calculated for Mito10-ATO C50H53ClO3P+ [M]+ 767.3415, found, 767.3420. 31P NMR (400.13 MHz, CDCl3) δ 24.48. 1H NMR (400.13 MHz, CDCl3, δ 8.08–7.98 (2H, m), 7.90–7.82 (6H, m), 7.81–7.75 (3H, m), 7.73–7.64 (8H, m), 7.25–7.23 (2H, m), 7.18–7.16 (2H, m), 4.30 (2H, t, J = 6.6), 3.88–3.77 (2H, m), 3.25–3.15 (1H, m), 2.66–2.55 (1H, m), 2.24–2.11 (2H, m), 2.01–1.92 (2H, m), 1.86–1.77 (2H, m), 1.76–1.68 (4H, m), 1.60–1.40 (6H, m), 1.38–1.25 (8H, m). 13C NMR (75 MHz, CDCl3) δ 185.5, 181.9, 158.0, 146.0, 138.6, 134.9, 134.8, 133.8, 133.72, 133.67, 133.1, 132.4, 131.5, 131.4, 130.5, 130.4, 128.4, 128.2, 126.3, 125.9, 118.9, 118.1, 74.0, 43.3, 35.4, 34.5, 30.4 (d, J = 14.0), 29.9, 29.5, 29.3, 29.2, 25.9, 22.8 (d, J = 49.1), 22.7 (d, J = 4.4).
NMR spectra and related parameters are given in Fig. S2.
Synthesis of Mito12-ATO
Mito12-ATO was prepared by reacting (12-bromodecyl)-triphenylphosphonium bromide with ATO in the presence of potassium carbonate in DMF (Fig. S1) as follows: (12-bromododecyl)-triphenylphosphonium bromide (0.6 g, 1.0 mmol) was added to a mixture of ATO (0.45 g, 1.2 mmol) and potassium carbonate (0.17 g, 1.2 mmol) in DMF. The mixture was stirred at 70 °C for 7 h. CH2Cl2 was added to the mixture followed by water (20 mL). The organic layer was washed twice with water and dried over Na2SO4. The solvent was removed under reduced pressure. Diethyl ether was then added to precipitate out the compound that was purified by flash chromatography (CH2Cl2/EtOH, 9:1) yielding the product, Mito12-ATO (0.35 g, 35% yield).
HRMS calculated for Mito12-ATO C52H57ClO3P+ [M]+ 795.3728, found, 795.3729.
31P NMR (400.13 MHz, CDCl3) δ 24.44. 1H NMR (400.13 MHz, CDCl3), δ 8.07–7.97 (2H, m), 7.87–7.74 (9H, m), 7.72–7.63 (8H, m), 7.24–7.20 (2H, m), 7.18–7.13 (2H, m), 4.30 (2H, t, J = 6.6), 3.82–3.72 (2H, m), 3.25–3.13 (1H, m), 2.65–2.55 (1H, m), 2.23–2.09 (2H, m), 1.99–1.92 (2H, m), 1.84–1.67 (7H, m), 1.63–1.42 (8H, m), 1.37–1.19 (9H, m). 13C NMR (75 MHz, CDCl3) δ 185.5, 181.9, 158.1, 146.0, 138.6, 134.92, 134.89, 133.7, 133.6, 133.1, 132.4, 131.5, 131.4, 130.5, 130.4, 128.4, 128.2, 126.3, 125.9, 118.9, 118.1, 74.1, 43.3, 35.4, 34.5, 30.5, 30.4, 30.3 (d, J = 16.1), 29.9, 29.6, 29.5, 29.3, 29.24, 29.21, 25.9, 22.8 (d, J = 49.2), 22.4 (d, J = 4.4).
The NMR spectra and related parameters are given in Fig. S2.
Synthesis of Mito16-ATO
Mito16-ATO was prepared by reacting (16-bromohexadecyl)-triphenylphosphonium bromide with ATO in the presence of potassium carbonate in DMF as follows: (16-bromohexadecyl)-triphenylphosphonium bromide (0.2 g, 0.34 mmol) was added to a mixture of ATO (0.13 g, 0.37 mmol) and potassium carbonate (0.05 g, 0.37 mmol) in DMF (2 mL). The mixture was stirred at 70 °C for 7 h. CH2Cl2 was added to the mixture as well as water (20 mL). The organic layer was washed twice with water and dried over Na2SO4. The solvent was removed under reduced pressure. Diethyl ether was the added to the mixture to precipitate out the compound that was purified by flash chromatography (CH2Cl2/EtOH, 9:1), yielding the product, Mito16-ATO (0.16 g, 54% yield).
HRMS calculated for Mito16-ATO C56H65ClO3P+ [M]+ 851.4354, found, 851.4360.
31P NMR (400.13 MHz, CDCl3) δ 23.39. 1H NMR (400.13 MHz, CDCl3), δ 8.00–7.93 (2H, m), 7.81–7.59 (17H, m), 7.21–7.16 (2H, m), 7.11–7.09 (2H, m), 4.26 (2H, t, J = 6.7), 3.75–3.63 (2H, m), 3.20–3.08 (1H, m), 2.60–2.48 (1H, m), 2.17–2.03 (2H, m), 1.94–1.85 (2H, m), 1.80–1.73 (2H, m), 1.70–1.63 (2H, m), 1.68–1.38 (8H, m), 1.33–1.06 (20H, m). 13C NMR (75 MHz, CDCl3) δ 185.54, 181.8, 158.0, 146.0, 138.5, 134.94, 134.91, 133.7, 133.6, 133.0, 132.3, 131.3, 131.4, 130.5, 130.4, 128.3, 128.1, 126.2, 125.9, 118.8, 118.0, 74.0, 43.3, 35.5, 34.5, 30.5, 30.4, 30.3, 29.9, 29.6, 29.59, 29.56, 29.52, 29.4, 29.3, 29.2, 29.1, 25.9, 22.8 (d, J = 49.9), 22.6 (d, J = 4.4).
The NMR spectra and related parameters are given in Fig. S2.
Cell culture
The following cell lines were obtained from the American Type Culture Collection (Manassas, VA), where they were regularly authenticated: MiaPaCa-2 (ATCC#CRL-1420, human pancreatic cancer), MDA-MB-231 (ATCC#HTB-26, human breast cancer), A549 (ATCC#CCL-185, human lung cancer), LLC (ATCC#CLR-1642, mouse lung cancer). LKR13 mouse lung cancer cells were a gift from Dr. Jonathan M. Kurie (MD Anderson Cancer Center, Houston, TX)30. All cell lines were grown at 37 °C in 5% CO2. MiaPaCa-2 and MDA-MB-231 cells were maintained in DMEM medium (Thermo Fisher Scientific, #11965) supplemented with 10% foetal bovine serum. A549, LLC, and LKR13 cells were maintained in RPMI 1640 medium (Thermo Fisher Scientific, #11875), supplemented with 10% foetal bovine serum. All cells were stored in liquid nitrogen and used within 20 passages after thawing.
Cell proliferation measurements
The IncuCyte Live-Cell Imaging system (IncuCyte Essen Bioscience Inc., Ann Arbor, MI) was used to monitor cell proliferation11–13. As shown in previous publications11,12, this imaging system is probe-free and noninvasive, and enables continuous monitoring of cell confluence over several days. The increase in the percentage of cell confluence was used as a surrogate marker of cell proliferation. In a 96-well plate, cells were plated at 1,000 cells per well in triplicates and left to adhere overnight. Cells were then treated with ATO, Mito-ATO analogs, and appropriate controls, and the cell confluency was recorded over several days in the IncuCyte S3 system.
Mitochondrial function measurements
Mitochondrial bioenergetic function was measured in real time using the Seahorse XF 96 Extracellular Flux Analyzer (Agilent, North Billerica, MA)11–13,22. The OCR-based assessment of mitochondrial complex activities was carried out on acutely permeabilized cells in the presence of different mitochondrial substrates, i.e., pyruvate/malate, for complex I and duroquinol for complex III11,14,31,32. Rot, malonate, and antimycin A (Sigma-Aldrich, St. Louis, MO) were used as specific inhibitors of mitochondrial complexes I, II, and III, respectively. Briefly, cells that were intact after treatments were immediately permeabilized using the Seahorse XF Plasma Membrane Permeabilizer (Agilent). The mitochondrial complex I-driven OCR was assayed in mannitol and sucrose buffer31 containing 10 mM pyruvate and 1.5 mM malate (substrates for complex I) and 10 mM malonate (which inhibits complex II activities). The mitochondrial complex III-driven OCR was assayed in mannitol and sucrose buffer containing 0.5 mM duroquinol (substrate for complex III) and 1 µM Rot and 10 mM malonate (which inhibit both complex I and II activities). The mitochondrial complex-dependent oxygen consumption (calculated as Rot- or antimycin A-inhibitable OCR equals basal OCR less OCR after Rot or antimycin A injection) was plotted against concentrations to determine the IC50 values.
Immunosuppression measurements
Mice
SMARTA triple reporter mice were generated in the following manner. First, IL-10 and IL-21 double-reporter mice33 were generated by cross-breeding IL-21tRFP mice34,35 with 10 BiT mice (kindly provided by Dr. Casey Weaver, University of Alabama at Birmingham). Double reporter mice were then crossed with GREAT (interferon-gamma reporter with endogenous polyA transcript) mice36 from Jackson Laboratory (Stock No. 017581). These triple-reporter mice were then crossed with SMARTA mice37 (kindly provided by Dr. Dorian McGavern, National Institutes of Health). Mice were bred and maintained in a closed breeding facility, and mouse handling conformed to the requirements of the Medical College of Wisconsin Institutional Animal Care and Use Committee guidelines. All experimental protocols were approved by the Medical College of Wisconsin Institutional Animal Care and Use Committee.
Cell culture
To differentiate CD4+ T cells into a T regulatory cell phenotype, splenocytes from SMARTA triple-reporter mice were processed and the red blood cell lysed using an ACK (ammonium-chloride-potassium) lysis buffer. The cells were then activated with 1 μg/mL GP61–80 peptide (GenScript, Piscataway, NJ) and 5 ng/mL TGF-β1 (Shenandoah Biotechnology, Inc., Warwick, PA). After one day of initial skewing, 100 μg/mL IL-2 along with ATO or Mito-ATO analogs of varying concentrations were added to the culture. Cells were cultured for six days and split once cells reached confluency; cells were replenished with IL-2 and compound accordingly. After six days in culture, cells were stained to assess the viability and phenotypic analysis via flow cytometry. LIVE/DEAD fixable violet or aqua dead cell stain (Invitrogen, Carlsbad, CA) was used to assess cell viability. The following antibodies were used for flow cytometry staining: PercP anti-mouse CD4 (clone: GK1.5; BioLegend, San Diego, CA), APC/Cy7 anti-mouse CD25 (clone: PC61; BioLegend), and PE anti-mouse FOXP3 (clone: FJK-16S; eBioscience, San Diego, CA). Flow cytometry data were acquired using a BD LSRII (BD Biosciences, CA) flow cytometer and analysed using FlowJo (Treestar, Inc., Ashland, OR).
Uptake of ATO and Mito10-ATO into MiaPaCa-2 and A549 cells
Intracellular levels of ATO and Mito10-ATO analogs were quantitated by LC–MS/MS11,12. Cells (1 × 106 per dish) were grown in 10-cm dishes and incubated with ATO and Mito10-ATO for 24 h in full culture media. The compounds were extracted according to the published experimental protocol11,12. Briefly, cells were washed twice with ice-cold DPBS (Dulbecco’s Phosphate Buffered Saline) and harvested. The cell pellet was immediately frozen in liquid nitrogen and stored at − 80 °C. For the extraction, the pellet was homogenized in 200 µl DPBS. A total of 2 µl were taken for the protein assay and 180 µl were extracted twice with a dichloromethane:methanol (2:1) mixture. The organic layers were combined and dried using a SpeedVac concentrator (Thermo Fisher Scientific, Waltham MA). The dry residue was dissolved in 100 µl of ice-cold methanol and taken for LC–MS/MS analysis. LC–MS/MS analyses were performed using a Kinetex Phenyl-Hexyl column (50 mm × 2.1 mm, 1.7 µm; Phenomenex, Torrance, CA) equilibrated with a water:acetonitrile mixture (4:1) containing 0.1% formic acid. Compounds were eluted by increasing the content of acetonitrile from 20 to 100% over 4 min and detected using the MRM (multiple reaction monitoring) mode. The protein concentrations were determined by the standard Bradford method (Bio-Rad, Hercules, CA). In brief, 2 µl of sample was added to 198 µl of preformulated Coomassie Brilliant Blue G-250 assay reagent (Bio-Rad), and the resultant blue colour was measured at 595 nm. The total protein level of the sample used to perform the extraction was calculated as milligrams of protein per sample. Intracellular concentrations (nmol/mg protein) of ATO and Mito10-ATO were calculated based on the LC–MS/MS standard curve, and the results were normalized to the total protein level.
Supplementary information
Acknowledgements
This work was supported in part by the Harry R. and Angeline E. Quadracci Professor in Parkinson’s Research Endowment (BK), the National Institutes of Health under grants AI125741 and AI148403 (WC) and CA246920 (TP), an American Cancer Society Research Scholar Grant (WC), and an Advancing a Healthier Wisconsin grant (WC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions
G.C., M.H., and B.K. designed the study. G.C. and M.H. developed the methodology. G.C., M.H., P.T., and R.Z. acquired the data (provided animals, provided facilities, etc.). G.C., M.H., and B.K. drafted the manuscript and created the figures. G.C., M.H., P.T., R.Z., P.V., W.C., and B.K. reviewed and revised the manuscript. G.C., M.H., and B.K. provided administrative, technical, and/or material support (i.e., reported or organized data, constructed databases). G.C. and BK supervised the study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-74808-0.
References
- 1.Fry M, Pudney M. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4′-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80) Biochem. Pharmacol. 1992;43:1545–1553. doi: 10.1016/0006-2952(92)90213-3. [DOI] [PubMed] [Google Scholar]
- 2.Araujo FG, Huskinson J, Remington JS. Remarkable in vitro and in vivo activities of the hydroxynaphthoquinone 566C80 against tachyzoites and tissue cysts of Toxoplasma gondii. Antimicrob. Agents Chemother. 1991;35:293–299. doi: 10.1128/aac.35.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cifuentes Kottkamp A, et al. Atovaquone inhibits arbovirus replication through the depletion of intracellular nucleotides. J. Virol. 2019 doi: 10.1128/jvi.00389-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kessl JJ, et al. Molecular basis for atovaquone binding to the cytochrome bc1 complex. J. Biol. Chem. 2003;278:31312–31318. doi: 10.1074/jbc.M304042200. [DOI] [PubMed] [Google Scholar]
- 5.Birth D, Kao W-C, Hunte C. Structural analysis of atovaquone-inhibited cytochrome bc1 complex reveals the molecular basis of antimalarial drug action. Nat. Commun. 2014;5:4029. doi: 10.1038/ncomms5029. [DOI] [PubMed] [Google Scholar]
- 6.Fiorillo M, et al. Repurposing atovaquone: Targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget. 2016;7:34084–34099. doi: 10.18632/oncotarget.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashton TM, et al. The anti-malarial atovaquone increases radiosensitivity by alleviating tumour hypoxia. Nat. Commun. 2016;7:12308. doi: 10.1038/ncomms12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nayak AP, Kapur A, Barroilhet L, Patankar MS. Oxidative phosphorylation: A target for novel therapeutic strategies against ovarian cancer. Cancers (Basel) 2018 doi: 10.3390/cancers10090337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang M, et al. Gene expression-based discovery of atovaquone as a STAT3 inhibitor and anticancer agent. Blood. 2016;128:1845–1853. doi: 10.1182/blood-2015-07-660506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takabe H, et al. A repurposed drug for brain cancer: Enhanced atovaquone amorphous solid dispersion by combining a spontaneously emulsifying component with a polymer carrier. Pharmaceutics. 2018 doi: 10.3390/pharmaceutics10020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng G, et al. Mitochondria-targeted analogues of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Cancer Res. 2016;76:3904–3915. doi: 10.1158/0008-5472.Can-15-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng G, et al. Mitochondria-targeted vitamin E analogs inhibit breast cancer cell energy metabolism and promote cell death. BMC Cancer. 2013;13:285. doi: 10.1186/1471-2407-13-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle KA, et al. Mitochondria-targeted drugs stimulate mitophagy and abrogate colon cancer cell proliferation. J. Biol. Chem. 2018;293:14891–14904. doi: 10.1074/jbc.RA117.001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng G, et al. Targeting lonidamine to mitochondria mitigates lung tumorigenesis and brain metastasis. Nat. Commun. 2019;10:2205. doi: 10.1038/s41467-019-10042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberg SE, et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zielonka J, et al. Mitochondria-targeted triphenylphosphonium-based compounds: Syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem. Rev. 2017;117:10043–10120. doi: 10.1021/acs.chemrev.7b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das M, Alzaid F, Bayry J. Regulatory T cells under the mercy of mitochondria. Cell Metab. 2019;29:243–245. doi: 10.1016/j.cmet.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Viswanadhan VN, Ghose AK, Revankar GR, Robins RK. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J. Chem. Inf. Comput. Sci. 1989;29:163–172. doi: 10.1021/ci00063a006. [DOI] [Google Scholar]
- 19.Klopman G, Li J-Y, Wang S, Dimayuga M. Computer automated log P calculations based on an extended group contribution approach. J. Chem. Inf. Comput. Sci. 1994;34:752–781. doi: 10.1021/ci00020a009. [DOI] [Google Scholar]
- 20.Barton V, Fisher N, Biagini GA, Ward SA, O'Neill PM. Inhibiting Plasmodium cytochrome bc1: A complex issue. Curr. Opin. Chem. Biol. 2010;14:440–446. doi: 10.1016/j.cbpa.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Sodero AC, et al. Insights into cytochrome bc1 complex binding mode of antimalarial 2-hydroxy-1,4-naphthoquinones through molecular modelling. Mem. Inst. Oswaldo Cruz. 2017;112:299–308. doi: 10.1590/0074-02760160417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ino Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer. 2013;108:914–923. doi: 10.1038/bjc.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang YA, et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol. Cancer. 2018;17:168. doi: 10.1186/s12943-018-0913-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, et al. Prognostic value of tumor-infiltrating Foxp3+ regulatory T cells in patients with breast cancer: a meta-analysis. J. Cancer. 2017;8:4098–4105. doi: 10.7150/jca.21030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhary B, Elkord E. Regulatory T cells in the tumor microenvironment and cancer progression: Role and therapeutic targeting. Vaccines (Basel) 2016;4:666. doi: 10.3390/vaccines4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohue Y, Nishikawa H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019;110:2080–2089. doi: 10.1111/cas.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shitara K, Nishikawa H. Regulatory T cells: A potential target in cancer immunotherapy. Ann. NY Acad. Sci. 2018;1417:104–115. doi: 10.1111/nyas.13625. [DOI] [PubMed] [Google Scholar]
- 29.Anso E, et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 2017;19:614–625. doi: 10.1038/ncb3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong L, et al. Identification of secreted proteins that mediate cell-cell interactions in an in vitro model of the lung cancer microenvironment. Cancer Res. 2008;68:7237–7245. doi: 10.1158/0008-5472.Can-08-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salabei JK, Gibb AA, Hill BG. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheaton WW, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xin G, et al. Single-cell RNA sequencing unveils an IL-10-producing helper subset that sustains humoral immunity during persistent infection. Nat. Commun. 2018;9:5037. doi: 10.1038/s41467-018-07492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xin G, et al. A critical role of IL-21-induced BATF in sustaining CD8-T-cell-mediated chronic viral control. Cell Rep. 2015;13:1118–1124. doi: 10.1016/j.celrep.2015.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinstein JS, et al. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 2016;17:1197–1205. doi: 10.1038/ni.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinhardt RL, Liang H-E, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-specific MHC-class II-restricted TCR-transgenic mice: Effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 1998;28:390–400. doi: 10.1002/(sici)1521-4141(199801)28:01<390::Aid-immu390>3.0.Co;2-o. [DOI] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.FlowJo Software v. 10 (Becton, Dickinson and Company, Ashland, OR, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.