Abstract
Background and Aim
About 15% patients with acute severe ulcerative colitis (UC) fail to respond to medical treatment and may require colectomy. An early prediction of response may help the treating team and the patients and their family to prepare for alternative treatment options.
Methods
Data of 263 patients (mean age 37.0 ± 14.0‐years, 176, 77% male) with acute severe UC admitted during a 12‐year period were used to study predictors of response using univariate analysis, multivariate linear principal component analysis (PCA), and nonlinear artificial neural network (ANN).
Results
Of 263 patients, 231 (87.8%) responded to the initial medical treatment that included oral prednisolone (n = 14, 5.3%), intravenous (IV) hydrocortisone (n = 238, 90.5%), IV cyclosporine (n = 9, 3.4%), and inflixmab (n = 2, 0.7%), and 28 (10.6%) did not respond and the remaining 4 (1.5%) died, all of whom did were also nonresponders. Nonresponding patients had to stay longer in the hospital and died more often. On univariate analysis, the presence of complications, the need for use of cyclosporin, lower Hb, platelets, albumin, serum potassium, and higher C‐reactive protein were predictors of nonresponse. Hb and albumin were strong predictive factors on both PCA and ANN. Though the nonlinear modeling using ANN had a good predictive accuracy for the response, its accuracy for predicting nonresponse was lower.
Conclusion
It is possible to predict the response to medical treatment in patients with UC using linear and nonlinear modeling technique. Serum albumin and Hb are strong predictive factors.
Keywords: artificial neural network, corticosteroid, deep learning, immunosuppressants, inflammatory bowel disease, medical treatment, outcome predictors
An early prediction of response of medical treatment, which occurs in 15% of patients with acute severe ulcerative colitis (UC) may help the treating team and the patients and their families to prepare for alternative treatment options. In this study on 263 patients with acute severe UC, we found that it is possible to predict the response to medical treatment in patients with UC using linear and non‐linear (artificial neural network) modeling techniques. Serum albumin and Hb are strong predictive factors.
Introduction
Inflammatory bowel disease, both ulcerative colitis (UC) and Crohn's disease are becoming increasingly common in India.1, 2 Despite recent advances in pharmacotherapy for UC, relapses of varying severity occur in 12–58% of patients with this disease while on treatment.3, 4, 5 Although the relapses of UC are quite effectively managed with intravenous corticosteroids currently, about 15% patients fail to respond to this form of treatment.5, 6, 7 Previously, these patients had to undergo colectomy during index hospitalization, which was associated with significant complications, morbidity, and mortality.8 Moreover, timing of surgery in acute severe colitis is still based on the physiciancs subjective assessment rather than on objective parameters.9 Currently, many such patients who fail to respond to 1‐week treatment with intravenous corticosteroids may be successfully managed with cyclosporine and biological.10, 11 However, an early prediction about which patients is likely to respond to initial intravenous corticosteroid and who will not, may help the treating team, the surgeons, and the patients and their family to prepare for an expensive and potentially hazardous therapeutic option including high‐grade immunosuppression.12
Some workers did evaluate a number of simple laboratory tests to determine the prognosis of patients with severe UC. However, most of these studies have assessed (i) only one or a few laboratory tests that are not widely available or (ii) a few clinical parameters in combination with one laboratory test, on small sample of patients with UC.13 We too previously evaluated a few parameters that might help to determine outcome of severe UC on a small number of patients. In that study on 55 patients, we found that some of the routinely available laboratory parameters such as hemoglobin (Hb) levels below 9 g/dL (sensitivity 80%, specificity 73%, area under the curve [AUC] 0.803), prothrombin time above 14 s (control 12; sensitivity 60%, specificity 75%, AUC 0.768), and serum C‐reactive protein (CRP) above 1.86 mg/dL (sensitivity 89%, specificity 59%, AUC 0.784) correctly categorized nonresponders.5 The present study was undertaken to evaluate parameters associated with failure of medical treatment among a large cohort of hospitalized patients with acute severe colitis to establish the validity of the above‐mentioned widely available laboratory parameters on a much larger sample of patients using the recently developed technologies such as principal component analysis (PCA)14 and artificial intelligence (artificial neural network, ANN).15
PCA is a statistical approach that reduces a set of interrelated variables into a few dimensions that gather a big amount of the variability of the original variables. These dimensions are called the components and have the properties of collecting highly correlated variables within each component and being uncorrelated with each other. The general rule is to select the principal components with the largest variance and keep only those that, explaining enough variance, make epidemiological and/or clinical sense.16 Whereas PCA is a linear method, ANN is a computerized mathematical nonlinear modeling technique, which includes a multilayer perceptron network consisting of multiple weighted regression equations in each layer that identifies the relationship between an unlimited number of input variables to predict the outcome variable.17, 18 Training of the ANN is done by data from a sample of patients whose outcome is known to the network and subsequently, its predictive accuracy is tested with another set of patients whose outcome is unknown to it. During training, the network adjusts the weight of the regression coefficients depending upon the degree of errors committed during prediction to obtain the highest degree of accuracy. However, in contrast to the other modeling techniques, ANN models never stops to learn. ANN is being widely used currently as a diagnostic and predictive modeling in several clinical and research settings.19
Methods
Patients
Prospectively maintained data of 263 patients with acute severe UC admitted to inpatient service of the Department of Gastroenterology of a multilevel teaching Institution during a 12‐year period (from 2000 to 2012) were used to train and test the predictive models. Those with incomplete records were excluded. Diagnosis of UC was based on clinical, colonoscopic, and histological parameters at the time of index admission or in the past or both, and exclusion of an infective cause by microbiological examination of stool.
Clinical parameters
At index admission, the parameters recorded included the age, sex, duration and extent of the disease (proctitis, left‐sided or pancolitis), duration of the index relapse and its severity (using Truelove–Witt's criteria and clinical activity score), extraintestinal manifestations (in skin, joints, and eyes), complications (e.g. toxic megacolon, shock), duration of hospitalization, treatment given, response to treatment, and the need for emergency colectomy.
Laboratory parameters
These included Hb (g/dL), total leukocyte count (TLC, 109/mm3), polymorphonuclear leukocyte count (%), platelet count (109/mm3), prothrombin time (test–control, seconds), albumin (g/dL), potassium (mmol/L), erythrocyte sedimentation rate (mm/h), and CRP (mg/dL).
Management and outcome
Each patient was managed as inpatient. Severity of the attack was assessed clinically using Truelove–Witts criteria. Oral (prednisolone) or intravenous glucocorticoides (hydrocortisone) was administered based on the severity of the attack on the discretion of the treating physician. Other treatment included broad‐spectrum antibiotics, and correction of fluid and electrolyte abnormalities. All the patients were allowed oral feed except in presence of toxic megacolon and/or paralytic ileus and suspicion of bowel perforation. Response was defined as reduction in clinical activity index (CAI) score20 below 10 for at least two consecutive days. In contrast, if the score did not reduce below 10 by day 7, the patient was considered as nonresponder. Moreover, worsening of the clinical condition while on treatment, development of complications such as life‐threatening gastrointestinal bleeding or perforation, and death during the same hospital admission were also recorded. Patients not responding to the initial 1 week treatment with intravenous hydrocortisone were treated with additional cyclosporine, infliximab, or surgery based on the joint decision by the medical and surgical gastroenterologists. Patients responding to initial intravenous hydrocortisone treatment were switched to equivalent dose of oral prednisolone and discharged on 5‐aminosalicylic acid. Oral prednisolone was tapered over the next 3–4 months. Steroid‐dependent patients as defined by relapse during tapering or within 1 month after discontinuation of the prednisolone and those requiring other immunosupressants in addition to hydrocortisone during the acute attack were treated with oral azathioprine (2 mg/kg of the body weight).
Building, training and testing of ANN
Construction of an ANN includes determining the network architecture, training, and testing the learned network. The feed‐forward multilayer perceptron network structure was constructed; signals traversed from 12 input (independent or predictor) variables through the three hidden units to predict the output (Fig. 1 ). The input (predictor) variable values were placed in the input units, which predicted the outcome variable through the hidden layers in sequential order. The activation value was calculated and passed through the activation function to produce the output. The back‐propagation algorithm was implemented to readjust the weights within the network to achieve the best possible predictive ability of the dichotomous outcome (response/no response) of the trained network. The intelligent problem solver toolbox was used to select the best network design. The 263 patients (responder 231, nonresponder 32) were randomly divided into two subsets, 132 for training the network, and the remaining for testing. If the classification confidence limit or the optimum threshold for minimum classification error was 0.95 then it was accepted and if it was found to be less than 0.05 then the values were rejected.
Figure 1.
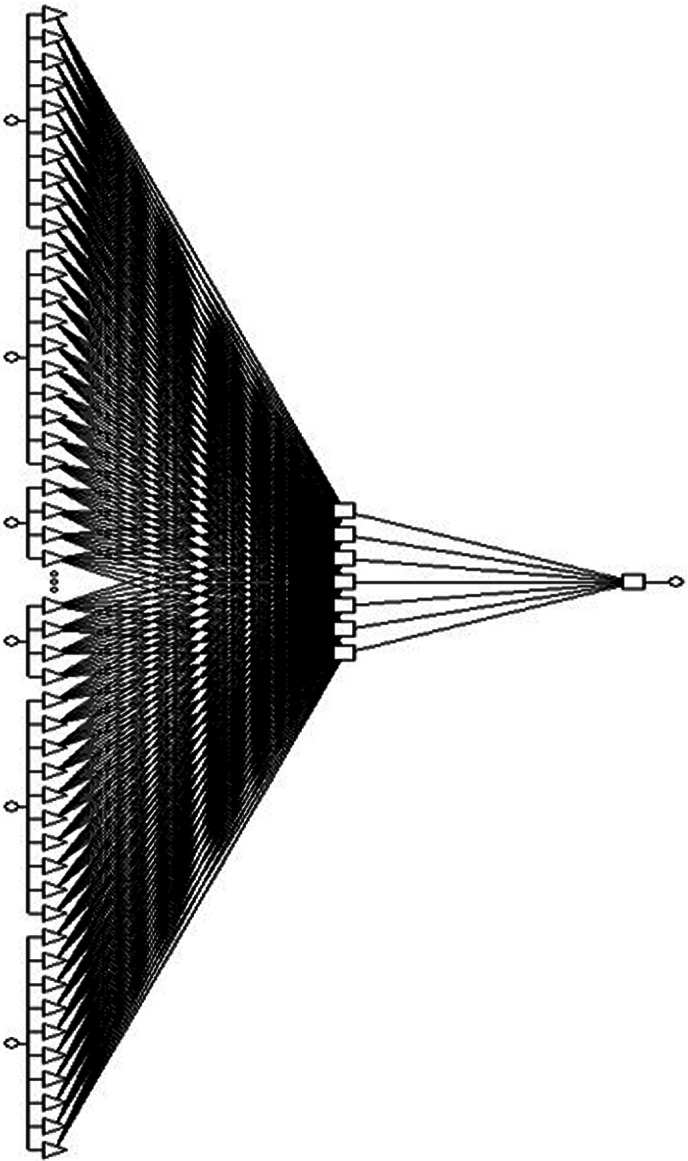
Schematic representation of the selected artificial neural network (ANN) model that includes 13 input variables, 3 hidden layers and single output variable.
The clinical and laboratory covariates (predictor variables) like age, sex, complications, TLC, differential count, platelet count, albumin, potassium, erythrocyte sedimentation rate (ESR), CRP, duration of hospital stay, and prothrombin time were used as the input variables. During training iterations, the network's predictive ability was corrected by the actual outcome; if the prediction was erroneous, the model re‐learned it by adjusting the weights and the hidden layers within the network during back‐propagation. All the statistical analyses were done using software (Statistica Neural Networks, Statsoft, Tulsa, USA), R, Epicalc and R‐studio (R development core team, Vienna, Austria), and MedCalc version 14 (Warandeberg 3, 1000 Brussels, Belgium).
Statistical analysis
Data were checked for distribution using Shapiro–Wilk test. Categorical data were presented as proportion. Parametric continuous data were presented as mean and standard deviation and nonparametric data as median and interquartile range. For univariate analysis, categorical variables were analyzed by chi‐squared test with Yates' correction, as applicable and parametric and nonparametric unpaired continuous data were analyzed by unpaired t test and Mann–Whitney U test, respectively. P values <0.05 were considered significant. The parameters found significant on univariate analysis were further evaluated by PCA. The ANN model was constructed using Statistica Neural Networks software (Statistica Neural Networks, Statsoft, Tulsa, USA) including all the variables.
Results
Patients
Of 263 consecutive patients (mean age 37.0 ± 14.0 years, 176, 77% male) with acute severe UC, most had chronic colitis (median duration of the disease before the index admission 24 months, range 1–360 months) and the mean duration of the current relapse was 33.6 days (range 2–120 days). Eleven (4.2%) patients had complications at admission, which included toxic megacolon in 7 (2.7%, 3 of whom had perforation as well), septic shock in 3 (1.1%), and perforation in 1 (0.4%). Extraintestinal manifestations included peripheral arthritis (n = 14, 5.3%), sacroilitis (n = 3, 1.1%), pyoderma gangrenosum (n = 5, 1.9%), primary sclerosing cholangitis (n = 1, 0.4%), deep venous thrombosis (n = 2, 0.8%, including one with cerebral cortical venous thrombosis), common carotid artery occlusion with cerebral infarction (n = 1, 0.4%, reported previously),21 oral ulcer (n = 2, 0.8%), skin rash (n = 2, 0.8%), scleritis (n = 1, 0.4%), and systemic amyloidosis (n = 1, 0.4%).
Laboratory parameters
The laboratory parameters at admission were as follows: Hb 9.5 ± 2.6 g/dL (normal 12–15.5), TLC 9.4 ± 5.2 × 109/mm3 (normal 4–11 × 109/mm3), polymorphs 75.9 ± 10.5%, platelets 263.8 ± 126.9 × 109/mm3(normal 150 000 to 450 000 × 109/mm3), serum albumin (median and range) 2.9 g/dL (1–6) (normal 3.5–5.5 g/dL), serum potassium 3.8 ± 0.6 mEq/L (normal 3.5–5.0), ESR 35.1 ± 17.2 mm/h (normal ≤15), and CRP (median and range) 1.7 mg/L (0.3–27.1) (normal ≤3 mg/L). Extent of the disease on colonoscopy was procosigmoiditis (n = 18, 6.8%), left‐sided colitis (n = 57, 21.7%), pancolitis (n = 95, 36.1%), and in 93 (35.4%) patients, data on extent were not available.
Outcome of treatment
Of 263 patients, 231 (87.8%) responded to the initial medical treatment that included oral prednisolone (n = 14, 5.3%), intravenous hydrocortisone (n = 238, 90.5%), intravenous cyclosporine added to intravenous hydrocortisone (n = 9, 3.4%), and infliximab (n = 2, 0.7%). Twenty‐eight (10.6%) did not respond and the remaining 4 (1.5%) patients died. All the patients who died did not respond clinically. Fourteen of the nonresponders underwent emergency colectomy and eight others left the hospital for not able to afford for surgical treatment.
Factors associated with nonresponse
Thirty‐two (12.1%) patients were nonresponders. Factors predicting response/nonresponse as binary variables were analyzed by univariate statistics, linear method using PCA, and nonlinear modeling by ANN.
Univariate analysis
Table 1 presents clinical and laboratory data of responders in comparison to nonresponders; presence of complications, need for use of cyclosporin in addition to corticosteroids, lower Hb, platelets, albumin, serum potassium, and higher CRP were associated with nonresponse (Fig. 2). Nonresponders had to stay in hospital longer than responders (median, interquartile range, 13 days, 8–19.8 vs 7 days, 5–10; P < 0.001) and died more often (4/32, 12.5% vs 0/231, 0%; P = <0.001, Table 1).
Table 1.
Demographic, clinical, and laboratory parameters of responsive and nonresponsive patients with ulcerative colitis
| Response (n = 231) | No response or death (n = 32) | P value | |
|---|---|---|---|
| Age (years, median, IQR) | 37 (25, 47) | 31 (24.8, 40.5) | 0.057 |
| Male gender | 155/231 (67.1%) | 21/32 (65.6%) | 0.973 |
| Duration of episode (days, median, IQR) | 30 (15, 45) | 30 (20, 45) | 0.317 |
| Treatment | |||
| Oral steroid | 14 (6.1) | 0 (0) | 0.004 |
| Intravenous (IV) steroids | 211 (91.3) | 27 (84.4) | |
| IV steroid + cyclosporine | 4 (1.7) | 5 (15.6) | |
| Infliximab | 2 (0.9) | 0 (0) | |
| Death | 0 (0) | 4 (12.5) | <0.001 |
| Complications | |||
| None | 229 (99.1) | 13 (59.1) | <0.001 |
| Toxic megacolon | 2 (0.9) | 2 (9.1) | |
| Megacolon + perforation | 0 (0) | 3 (13.6) | |
| Septic shock | 0 (0) | 3 (13.6) | |
| Perforatopn | 0 (0) | 1 (4.5) | |
| Hb (g/dL, mean, SD) | 9.6 (2.6) | 8.6 (2) | 0.044 |
| Total leukocyte count (×109/mm3, median, IQR) | 8.4 (6.5, 11.1) | 7.9 (6, 10.9) | 0.516 |
| Polymorphonuclear leukocyte (%, median, IQR) | 76.5 (70, 84) | 78.5 (71.5, 84) | 0.455 |
| Platelet (×109/mm3, median, IQR) | 256 (195, 327) | 189 (104.5, 289.5) | 0.009 |
| Albumin (median, IQR) | 3 (2.3, 3.5) | 2.3 (1.9, 2.6) | <0.001 |
| Potassium (median, IQR) | 3.9 (3.5, 4.3) | 3.6 (3.2, 4.1) | 0.023 |
| Erythrocyte sedimentation rate (mm/h, median, IQR) | 35 (20, 48) | 41 (23, 56.8) | 0.331 |
| CRP (mg/L, median, IQR) | 1.6 (0.5, 4.1) | 6 (2.1, 9.7) | 0.002 |
| Treatment with cyclosporine or infliximab | 6/231 (2.6%) | 5/32 (15.6%) | 0.005 |
| Duration of hospital stay (days, median, IQR) | 7 (5, 10) | 13 (8, 19.8) | <0.001 |
CRP, serum C‐reactive protein; Hb, hemoglobin; n, number; IQR, interquartile range.
Figure 2.
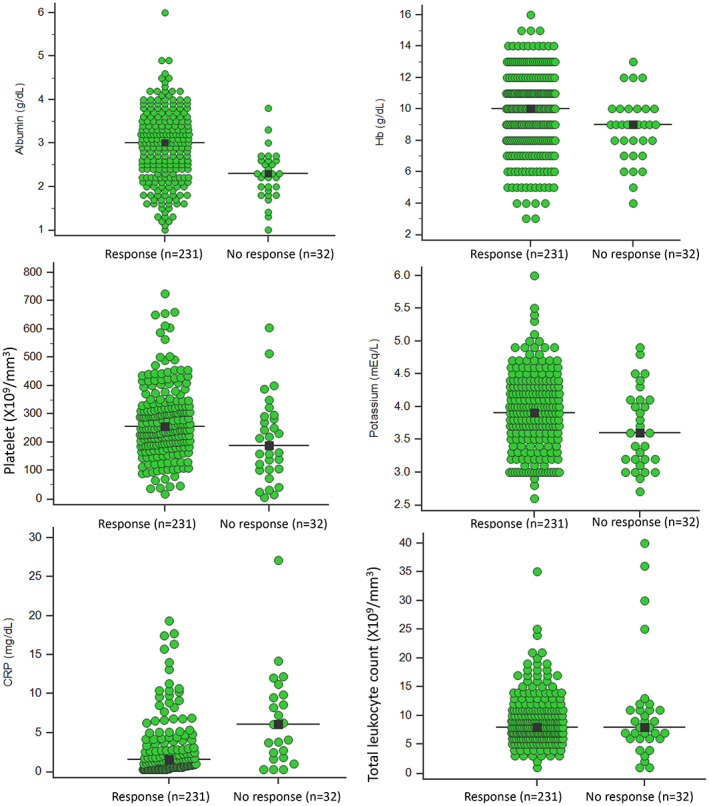
Univariate analysis of clinical and laboratory variables of responders in comparison to nonresponders.
Principal component analysis
PCA of the laboratory parameters significant on univariate analysis, namely TLC, Hb, albumin, K, platelet, and CRP, the first three principle components showed higher eigenvalues (measure the variance of the explanatory “strength” of principal components) with large cumulative variance of 66.3% (Table 2). However, discriminative ability of the parameters analyzed was not very high as there was reasonable degree of overlap (Fig. 3). In our study, Hb, and albumin contributed maximum ability as measures of predictive factors (Fig. 4, Table 3).
Table 2.
The proportion of variance explained by each principal component in the data set
| Principal components | Eigenvalues | Variance (%) | Cumulative variance (%) |
|---|---|---|---|
| Parameters: TLC, Hb, albumin | |||
| PC 1 | 1.48 | 49.42 | 49.41 |
| PC 2 | 0.92 | 30.51 | 79.93 |
| PC 3 | 0.60 | 20.07 | 100 |
| Parameters: TLC, Hb, albumin, K | |||
| PC 1 | 1.55 | 38.65 | 38.65 |
| PC 2 | 0.96 | 23.98 | 62.63 |
| PC 3 | 0.92 | 22.88 | 85.51 |
| PC 4 | 0.58 | 14.49 | 100 |
| Parameters: TLC, Hb, albumin, K, platelet | |||
| PC 1 | 1.61 | 32.23 | 32.23 |
| PC 2 | 1.25 | 24.99 | 57.22 |
| PC 3 | 0.93 | 18.66 | 75.87 |
| PC 4 | 0.72 | 14.49 | 90.37 |
| PC 5 | 0.48 | 9.63 | 100 |
| Parameters: TLC, Hb, albumin, K, platelet, CRP | |||
| PC 1 | 1.67 | 27.92 | 27.92 |
| PC 2 | 1.25 | 20.86 | 48.79 |
| PC 3 | 1.05 | 17.48 | 66.27 |
| PC 4 | 0.83 | 13.87 | 80.14 |
| PC 5 | 0.69 | 11.62 | 91.76 |
| PC 6 | 0.49 | 8.24 | 100 |
CRP, serum C‐reactive protein; Hb, hemoglobin; K, potassium; PC, principal component; TLC, total leukocyte count.
Figure 3.
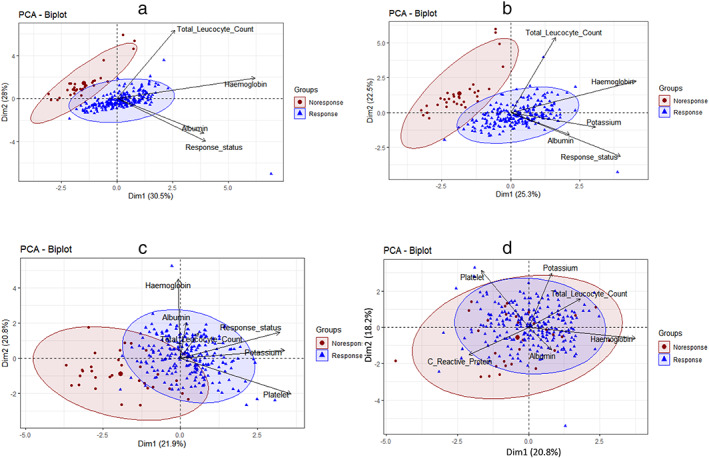
Principal component biplots of significant parameters (total leukocyte count, hemoglobin, albumin, potassium, platelet, C‐reactive protein) of responders and nonresponders. Biplots of contributing variables show distinct separation of outcome variables. (a) biplot of three variables total leukocyte count, hemoglobin, and albumin; (b) biplot of four variables total leukocyte count, hemoglobin, albumin and potassium; (c) biplot of five variables total leukocyte count, hemoglobin, albumin, potassium, and platelet; (d) biplot of four variables total leukocyte count, hemoglobin, albumin, potassium, and C‐reactive protein.  , Nonresponse;
, Nonresponse;  , response.
, response.
Figure 4.
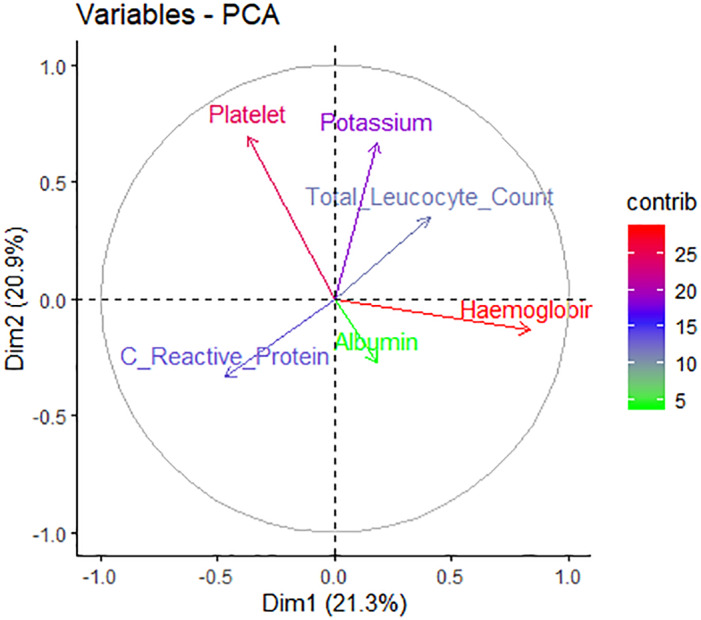
Contribution of different variables on principal component analysis (PCA) in two‐dimensions (dim).
Table 3.
Contribution of variables on each principal component
| Parameters | PC 1 | PC 2 |
|---|---|---|
| Total leukocyte count | 17.34 | 3.82 |
| Hemoglobin | 19.79 | 28.89 |
| Albumin | 28.00 | 10.44 |
| Potassium | 15.60 | 13.14 |
| Platelet | 9.96 | 43.19 |
| Serum C‐reactive protein | 9.30 | 0.51 |
PC, principal component.
ANN model
To predict the response to medical treatment in UC patients, the multilayer perceptron neural network was trained by back‐propagation algorithm (10 networks retained out of 16 tested). The classification accuracy rate was 73% in correctly classifying response to medical treatment in UC patients. While testing, the network performed well for predicting response as an outcome variable though it was not as good for predicting nonresponse (Fig. 1 ). The distribution of the generalized weights showed covariate age had no effect on the outcome since the distribution of generalized weights was nearly zero and that at least the two covariates Hb and albumin had a nonlinear effect since the variance of their generalized weights were overall greater than one (Fig. 5 ). The most powerful predictors of response outcome were Hb, albumin and platelet count and this result was in concordance with the results of PCA and univariate analysis.
Figure 5.
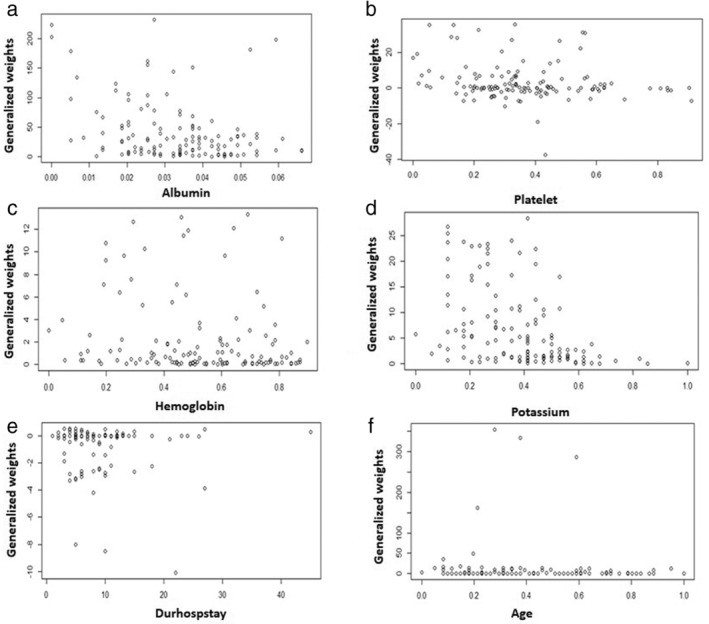
Plots of generalized weights with respect to each covariate.
Discussion
The current single‐center study on a large cohort of patients with acute severe UC shows that (i) about 12% of such patients do not respond to initial medical treatment, (ii) on univariate analysis, presence of complications, need for use of cyclosporin in addition to corticosteroids, lower Hb, platelet count, albumin, serum potassium levels, and higher CRP were predictors of nonresponse, (iii) nonresponding patients had to stay longer in hospital and died more often, (iv) Hb and albumin contributed maximum as predictive factors on PCA, and (v) though ANN take all the predictive variables into consideration, Hb and albumin had the strongest predictive influence, which was similar to the results of PCA.
The results of our study showing 12% nonresponse among patients with acute severe UC, though somewhat close to the previous literature but was somewhat lower.5, 22, 23, 24, 25, 26 This might be related to the fact that our patients are generally referred from other hospitals and hence, are more likely to have severe disease; moreover, compared to some of the earlier studies, the number of patients included in the current study was quite large. In a previous study from northern India on 179 patients with acute severe colitis evaluating long‐term predictors of response using machine learning algorithms or artificial intelligence, 19 (11%) patients underwent colectomy at index admission.27 Authors found that response at day 7 of hospitalization, steroid use during first year of diagnosis, longer disease duration prior to acute severe colitis and number of extraintestinal manifestations, were able to predict colectomy with an accuracy of 77%. In our series, 14/263 (5.3%) underwent colectomy, which is lower than that study; however, 8 (3%) other nonresponding patients left hospital as they could not afford for surgical treatment.
Results of the univariate analysis, linear multivariate analysis using PCA and nonlinear modeling using ANN consistently showed that a few parameters uniformly contributed toward the outcome predication, which is in accordance with our earlier study on a small sample of 55 patients. These parameters included serum albumin, Hb and platelet counts. These results are of clinical importance to predict nonresponse in patients admitted to hospital with acute severe UC. Importance of these parameters cannot be overestimated not only because of their predictive value but also because these are widely available laboratory tests even in peripheral hospitals. An early prediction or response to medical treatment may help the treating team, the surgeons, and the patients and their family members to prepare for an expensive and potentially hazardous therapeutic option including high‐grade immunosuppression. Hence, we believe that the result of the current study is of reasonable clinical importance.
Strong predictive ability of low Hb albumin might be related to the fact that these are surrogate markers of more severe disease. In a Swedish study on 171 patients with UC, extensive disease (hazard ratio = 2.40; confidence interval: 1.10–5.36) was found to be associated with an increased risk of anemia.28 In a Spanish study on 136 patients with UC, an older age at diagnosis, longer interval from diagnosis to corticosteroid‐therapy, lower CRP and higher Hb predicted better prognosis.29 In our previous study on 55 patients, low Hb and serum albumin levels were associated with nonresponse to medical treatment,5 which is accordance with the findings of the present study on a larger sample of patients.
Though the current ANN model predicted response with reasonable degree of accuracy, its predictive ability for the nonresponse was somewhat lesser. This is not unexpected considering a small number of patients in nonresponsive subgroup. However, ANN models have an advantage that it never stops learning and continues to improve its ability to predict with inclusion of more patients. Another advantage of ANN is inclusion of all the variables as has been done in current analysis rather than only a few selected variables found significant on univariate analysis. Such an approach makes ANN superior over conventional analysis as in conventional analysis, the data found significant on univariate analysis are only included in multivariate modeling. Since some of the data may not reach statistical significance on univariate analysis due to smaller number of patients having such co‐morbidity. For example, a patient with acute relapse of UC with cytomegalovirus infection with uncontrolled diabetes mellitus is more likely to die than if he/she did not have the latter two associated conditions30; however, due to the fact that a small number of patients may have such associations, these may not reach statistical significance during univariate analysis. Retrospective nature of the data is a limitation of the study. Another limitation of the current ANN model is lack of external validation, which is needed to improve its robustness.15 Moreover, it should be further validated in a prospectively included cohort of patients.
Artificial intelligence, also known as machine learning, is a nonlinear mathematical modeling technology that is used extensively in modern day living, such as email communications, social media, web searching, stores and services, banking and finance, aviation and prediction of machinery failure, maps and directions, criminology, and war.15, 31 Recently, artificial intelligence is being increasingly used in clinical medicine including gastroenterology32, 33 endoscopy,34 and hepatology,15 radiology,35 pathology,36 dentistry,37 oncology,38 cardiology,39 dermatology,40 neurosurgery,41 gynecology,42 and in medical research, particularly big data analysis. Whereas convolutional neural network is the usual network used for image analysis,43 feed‐forward multilayer perceptron networks are the modeling technique for clinical prediction and have been used in the current study as well. Particular advantages of artificial intelligence, that place this technology potentially in higher position than the other modeling techniques include nonlinear method of data analysis, ability to continue learning like human brain by back‐propagation and autocorrection, and inclusion of all the variables for prediction rather than a limited number of parameters. However, limitation of this technology is lack of familiarity among clinical personnel, and over‐learning or over‐fit by the models that reduce their broader utility or robustness. However, these limitations are being overcome by inclusion of such networks into machines that automatically help the clinicians in the diagnosis without the need of their familiarity with the technology itself. Though such “black box” approach has its own limitations, it is likely to result in broader application of this technology in different branches of medicine in future.
In conclusion, the prediction of response to medical treatment in patients with UC using linear and nonlinear modeling technique is possible taking serum albumin and Hb into consideration. Therefore, prospective clinical studies will be needed to evaluate the use of such models in decision‐making.
References
- 1. Philip M, Augustine P, Thomas V et al Multi‐center prospective survey of inflammatory bowel diseases in Kerala: more than 2000 cases. Indian J Gastroenterol. 2017; 36: 459–67. [DOI] [PubMed] [Google Scholar]
- 2. Makharia GK, Ramakrishna BS, Abraham P et al Survey of inflammatory bowel diseases in India. Indian J Gastroenterol. 2012; 31: 299–306. [DOI] [PubMed] [Google Scholar]
- 3. Ramakrishna BS, Makharia GK, Abraham P et al Indian Society of Gastroenterology consensus on ulcerative colitis. Indian J. Gastroenterol. 2012; 31: 307–23. [DOI] [PubMed] [Google Scholar]
- 4. Williams HR, Willsmore JD, Cox IJ et al Serum metabolic profiling in inflammatory bowel disease. Dig. Dis. Sci. 2012; 57: 2157–65. [DOI] [PubMed] [Google Scholar]
- 5. Kumar S, Ghoshal UC, Aggarwal R, Saraswat VA, Choudhuri G. Severe ulcerative colitis: prospective study of parameters determining outcome. J. Gastroenterol. Hepatol. 2004; 19: 1247–52. [DOI] [PubMed] [Google Scholar]
- 6. Kedia S, Ahuja V, Tandon R. Management of acute severe ulcerative colitis. World J Gastrointest Pathophysiol. 2014; 5: 579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharma MP, Makharia G. In search of a better treatment for ulcerative colitis. Indian J. Gastroenterol. 2003; 22: 77–8. [PubMed] [Google Scholar]
- 8. Mokhele NN, Thomson SR, Watermeyer GA. Predictors of emergency colectomy in patients admitted with acute severe ulcerative colitis. S. Afr. J. Surg. 2017; 55: 20–6. [PubMed] [Google Scholar]
- 9. Brahmania M, Bernstein CN. Physician global assessments or blood tests do not predict mucosal healing in ulcerative colitis. Can. J. Gastroenterol. Hepatol. 2014; 28: 325–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghoshal UC, Verma A. Biologicals in treatment of acute ulcerative colitis. Trop. Gastroenterol. 2015; 36: 80–5. [DOI] [PubMed] [Google Scholar]
- 11. Kamat N, Kedia S, Ghoshal UC et al Effectiveness and safety of adalimumab biosimilar in inflammatory bowel disease: a multicenter study. Indian J Gastroenterol. 2019; 38: 44–54. [DOI] [PubMed] [Google Scholar]
- 12. Sood A, Midha V, Sharma S et al Infliximab in patients with severe steroid‐refractory ulcerative colitis: Indian experience. Indian J. Gastroenterol. 2014; 33: 31–4. [DOI] [PubMed] [Google Scholar]
- 13. Cioffi M, Rosa AD, Serao R, Picone I, Vietri MT. Laboratory markers in ulcerative colitis: current insights and future advances. World J Gastrointest Pathophysiol. 2015; 6: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hotte NS, Salim SY, Tso RH et al Patients with inflammatory bowel disease exhibit dysregulated responses to microbial DNA. PLoS One. 2012; 7: e37932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banerjee R, Das A, Ghoshal UC, Sinha M. Predicting mortality in patients with cirrhosis of liver with application of neural network technology. J. Gastroenterol. Hepatol. 2003; 18: 1054–60. [DOI] [PubMed] [Google Scholar]
- 16. Zhang Z, Castello A. Principal components analysis in clinical studies. Ann Transl Med. 2017; 5: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cross SS, Harrison RF, Kennedy RL. Introduction to neural networks. Lancet. 1995; 346: 1075–9. [DOI] [PubMed] [Google Scholar]
- 18. Ghoshal UC, Das A. Models for prediction of mortality from cirrhosis with special reference to artificial neural network: a critical review. Hepatol Int. 2008; 2: 31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baxt WG. Application of artificial neural networks to clinical medicine. Lancet. 1995; 346: 1135–8. [DOI] [PubMed] [Google Scholar]
- 20. Lichtiger S, Present DH, Kornbluth A et al Cyclosporine in severe ulcerative colitis refractory to steroid therapy. N. Engl. J. Med. 1994; 330: 1841–5. [DOI] [PubMed] [Google Scholar]
- 21. Vavricka SR, Brun L, Ballabeni P et al Frequency and risk factors for extraintestinal manifestations in the Swiss inflammatory bowel disease cohort. Am. J. Gastroenterol. 2011; 106: 110–19. [DOI] [PubMed] [Google Scholar]
- 22. Chakravarty BJ. Predictors and the rate of medical treatment failure in ulcerative colitis. Am. J. Gastroenterol. 1993; 88: 852–5. [PubMed] [Google Scholar]
- 23. Truelove SC, Willoughby CP, Lee EG, Kettlewell MG. Further experience in the treatment of severe attacks of ulcerative colitis. Lancet. 1978; 2: 1086–8. [DOI] [PubMed] [Google Scholar]
- 24. Lennard‐Jones JE, Ritchie JK, Hilder W, Spicer CC. Assessment of severity in colitis: a preliminary study. Gut. 1975; 16: 579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oshitani N, Kitano A, Fukushima R et al Predictive factors for the response of ulcerative colitis patients during the acute‐phase treatment. Digestion. 1990; 46: 107–13. [DOI] [PubMed] [Google Scholar]
- 26. Chetri K, Ghoshal UC, Somani SK et al Common carotid artery occlusion causing cerebral infarction in ulcerative colitis. Indian J. Gastroenterol. 2002; 21: 122–3. [PubMed] [Google Scholar]
- 27. Jain S, Kedia S, Sethi T et al Predictors of long‐term outcomes in patients with acute severe colitis: a northern Indian cohort study. J. Gastroenterol. Hepatol. 2018; 33: 615–22. [DOI] [PubMed] [Google Scholar]
- 28. Eriksson C, Henriksson I, Brus O et al Incidence, prevalence and clinical outcome of anaemia in inflammatory bowel disease: a population‐based cohort study. Aliment. Pharmacol. Ther. 2018; 48: 638–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barreiro‐Alonso E, Saro‐Gismera C, Sanchez M. Outcomes and prediction of corticosteroid therapy after successive courses of ulcerative colitis treatments. Expert Rev. Gastroenterol. Hepatol. 2018; 12: 733–41. [DOI] [PubMed] [Google Scholar]
- 30. Kishore J, Ghoshal U, Ghoshal UC et al Infection with cytomegalovirus in patients with inflammatory bowel disease: prevalence, clinical significance and outcome. J. Med. Microbiol. 2004; 53: 1155–60. [DOI] [PubMed] [Google Scholar]
- 31. Chan JL, Purohit H. Challenges to transforming unconventional social media data into actionable knowledge for public health systems during disasters. Disaster Med. Public Health Prep. 2019: 1–8. [DOI] [PubMed] [Google Scholar]
- 32. Das A, Ben‐Menachem T, Cooper GS et al Prediction of outcome in acute lower‐gastrointestinal haemorrhage based on an artificial neural network: internal and external validation of a predictive model. Lancet. 2003; 362: 1261–6. [DOI] [PubMed] [Google Scholar]
- 33. Le Berre C, Sandborn WJ, Aridhi S et al Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology. 2020; 158: 76–94.e2. [DOI] [PubMed] [Google Scholar]
- 34. Hassan C, Wallace MB, Sharma P et al New artificial intelligence system: first validation study versus experienced endoscopists for colorectal polyp detection. Gut. 2019: 1. [DOI] [PubMed] [Google Scholar]
- 35. Qin ZZ, Sander MS. B R: using artificial intelligence to read chest radiographs for tuberculosis detection: a multi‐site evaluation of the diagnostic accuracy of three deep learning systems. Sci Rep. 2019; 6: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Serag A, Ion‐Margineanu A, Qureshi H et al Translational AI and deep learning in diagnostic pathology. Front. Med. 2019; 6: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hwang JJ, Jung YH, Cho BH, Heo MS. An overview of deep learning in the field of dentistry. Imaging Sci Dent. 2019; 49: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar SS, Awan KH, Patil S, Sk IB, Raj AT. Potential role of machine learning in oncology. J. Contemp. Dent. Pract. 2019; 20: 529–30. [PubMed] [Google Scholar]
- 39. Zhang Y, van der Werf NR, Jiang B, van Hamersvelt R, Greuter MJ, Xie X. Motion‐corrected coronary calcium scores by a convolutional neural network: a robotic simulating study. Eur. Radiol. 2020; 30: 1285–1294. [DOI] [PubMed] [Google Scholar]
- 40. Thomsen K, Iversen L, Titlestad TL, Winther O. Systematic review of machine learning for diagnosis and prognosis in dermatology. J. Dermatolog. Treat. 2019: 1–15. [DOI] [PubMed] [Google Scholar]
- 41. Sotoudeh H, Shafaat O, Bernstock JD et al Artificial intelligence in the management of glioma: era of personalized medicine. Front. Oncol. 2019; 9: 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moawad G, Tyan P, Louie M. Artificial intelligence and augmented reality in gynecology. Curr. Opin. Obstet. Gynecol. 2019; 31: 345–8. [DOI] [PubMed] [Google Scholar]
- 43. Horiuchi Y, Aoyama K, Tokai Y et al Convolutional neural network for differentiating gastric cancer from gastritis using magnified endoscopy with narrow band imaging. Dig. Dis. Sci. 2019; 10. [DOI] [PubMed] [Google Scholar]


