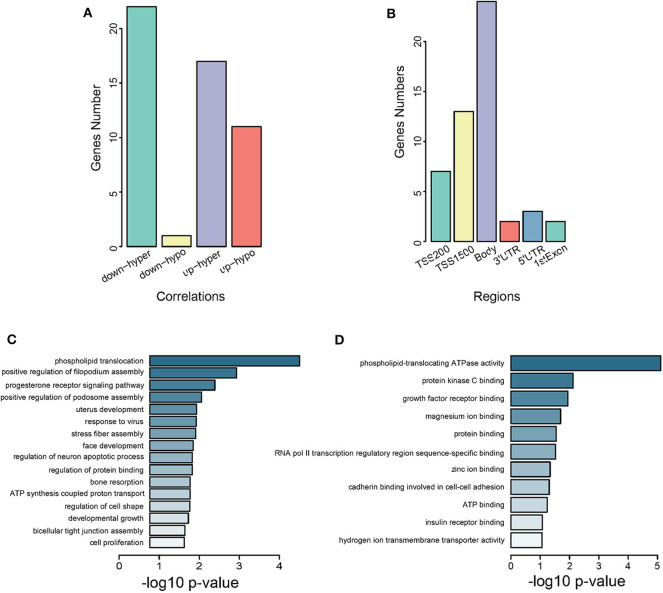
Figure 3.
Characterization of intersected DMGs. (A) Grouping of intersected genes by methylation levels. Down-hyper represents downregulated hyper-methylated genes. Up-hypo represents upregulated hypo-methylated genes. Up-hyper represents upregulated hyper-methylated genes. Down-hypo represents downregulated hypo-methylated genes. The y-axis is the number of genes and the x-axis indicates different groups. (B) Grouping of intersected genes by CpG methylation regions. The y-axis is the number of intersected genes. The x-axis labels different gene regions: TSS1500, TSS200, 5′UTR, 1stExon, body, and 3′UTR. (C) Subontology of biological process (BP) for intersected genes. (D) Subontology of molecular function (MF) for intersected genes. (C,D) The x-axis indicates the log p-value. The y-axis is the enriched terms.