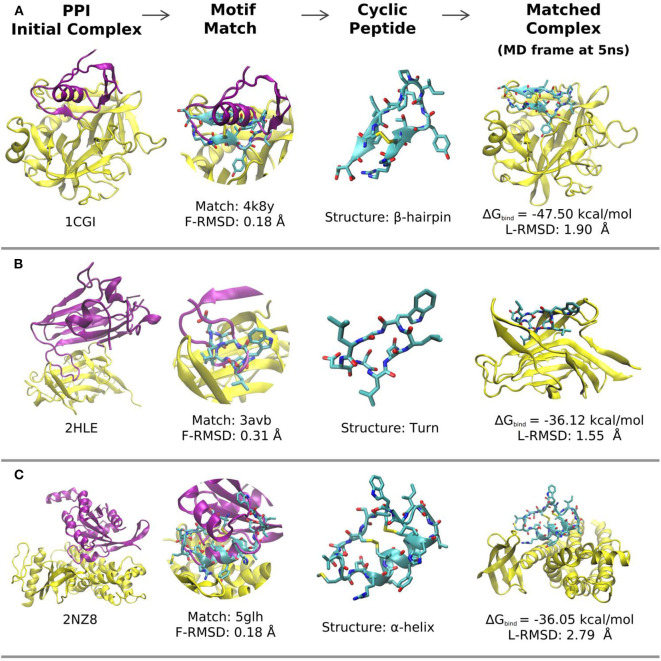
Figure 3.
Representative matches and modeled structures of protein-cyclic-peptide complexes. (A) Example of a cyclic peptide with an β-hairpin motif at the interface (pdb-entries of the complex and the cyclic peptide template are indicated; the calculated MMGBSA interaction energy and final deviation from the start structure are also included). (B) Same as in (A) but for a cyclic peptide with a turn motif as binder. (C) Same as in (A,B) for a cyclic α-helix binding motif. In each case the target protein-protein complex is shown as cartoon (yellow: receptor; pink: ligand protein). The superimposed matching cyclic peptide is indicated in the second column and the cyclic peptide (with adapted interface sequence) is shown in the third column. The last column represents the final structures of the receptor protein (yellow) in complex with the cyclic peptide after 5 ns MD simulation in explicit solvent.