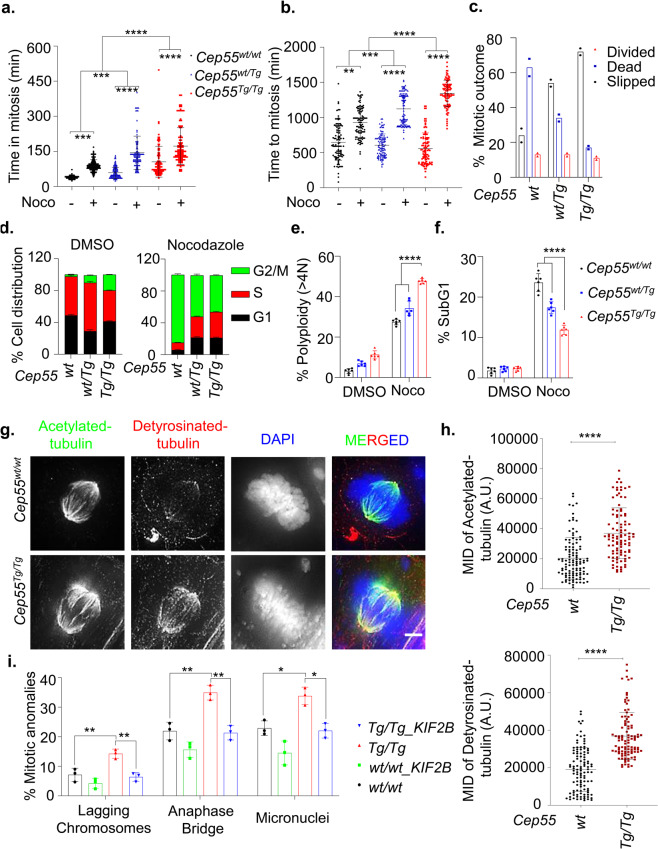
Fig. 7. Association of CEP55 with microtubule stability.
a, b Comparison of average time spent in mitosis (a) and average time taken to complete mitosis (b) determined using time-lapse microscopy of the immortalized MEFs of indicated genotypes in the presence and absence of nocodazole (0.5 μM). Error bars represent the ± SD from two independent experiments (fate of n = 50 cell were counted per experiment). Student’s t test was performed to determine P-value < 0.05 (*), < 0.01 (**), < 0.001 (***) and < 0.0001 (****). c Percentage of mitotic outcome of the immortalized MEFs of indicated genotypes in presence of nocodazole (0.5 μM) as shown in panel A and B. Mitotic slippage was defined by premature mitotic exit during nocodazole-induced mitotic arrest, while death was determined through membrane blebbing. Mean derived from two independent experiments (fate of n = 50 cell were counted per experiment). d Cell-cycle profiles of immortalized MEFs of indicated genotype in the presence or absence of nocodazole (0.5 μM) determined using FACS. Error bars represent the ± SD from three independent experiments. Two-way ANOVA test was performed to determine P-value as demonstrated in Supplementary Table 3. e, f Polyploidy (>4N DNA content) (e) and percentage of SubG1 populations (f) determined using FACS in the indicated immortalized MEFs in the presence or absence of nocodazole (0.5 μM). Error bars represent the ± SD from three independent experiments with two replicates each. One-way ANOVA test was performed to determine P-value < 0.0001 (****). g Representative images of detryosinated (red) and acetylated tubulin (green) of metaphase stages of immortalized Cep55wt/wt and Cep55Tg/Tg MEFs showing the microtubule networks (Scale bar, 100 μm). h Quantification of the mean integrated density (MID) of Acetylated (Upper panel) and detyrosinated (lower panel) tubulin observed among immortalized Cep55wt/wt and Cep55Tg/Tg MEFs. The intensity was calculated using Image J software wherein n = 50 metaphase cells were calculated per genotype. Error bars represent the ± SD from two independent experiments. Student’s t test was performed to determine P-value < 0.0001 (****). i Percentage of mitotic defects (described previously in Fig. 6f upon KIF2B overexpression in the immortalized MEFs (n = 100 cell per experiment were counted) of indicated genotypes. Error bars represent the ± SD from three independent experiments. One-way ANOVA test was performed to determine P-value < 0.05 (*) and < 0.01 (**).