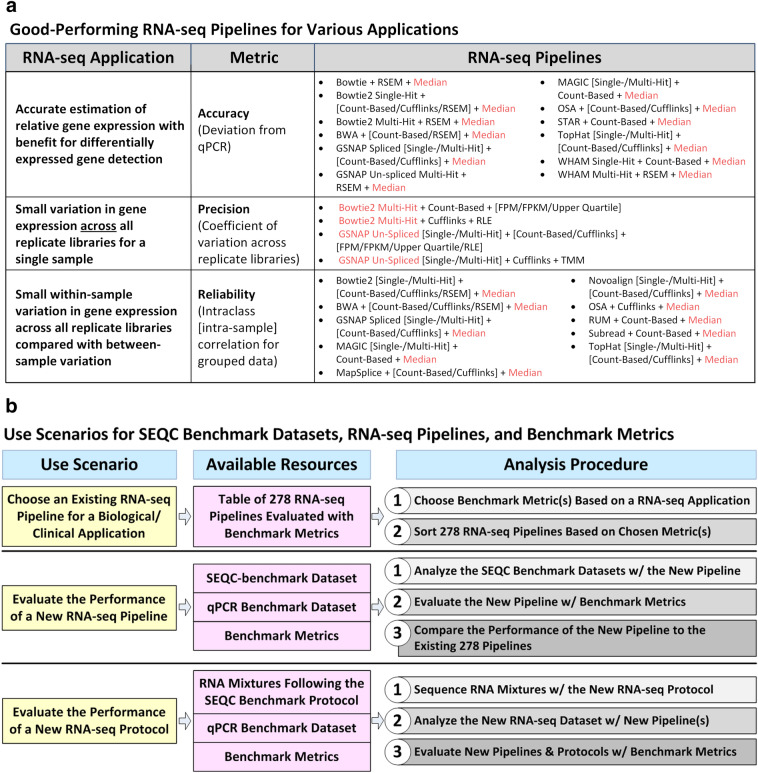
Figure 6.
The resources provided by this study (i.e., the 278 RNA-seq pipelines, the benchmark metrics, and the SEQC-benchmark datasets) can serve as guidelines for biological and clinical researchers as well as for bioinformaticians and biotechnologists. (a) Depending on the gene expression application, the three metrics (i.e., accuracy, precision, and reliability) may be used to choose a pipeline. We have associated each metric with an RNA-seq application and listed the top-performing pipelines for each metric. The red-highlighted component in each listed RNA-seq pipeline indicates components that occur frequently among the top-performing pipelines for each metric. (b) Biological or clinical researchers who want to analyze Illumina RNA-seq data (or data from similar platforms with short, fixed-length reads) can choose an existing RNA-seq pipeline using the provided table of 278 pipelines ranked by accuracy, precision, or reliability. Bioinformaticians that are developing a new RNA-seq pipeline for Illumina data (or data from similar platforms) can use the SEQC-benchmark datasets and benchmark metrics to evaluate the new pipeline and assess its performance relative to the 278 pipelines. Bioinformaticians or biotechnologists that are developing new RNA-seq protocols can first sequence the same RNA mixture samples (i.e., samples A, B, C, and D), and then evaluate associated data analysis pipelines using the qPCR benchmark dataset and the benchmark metrics.