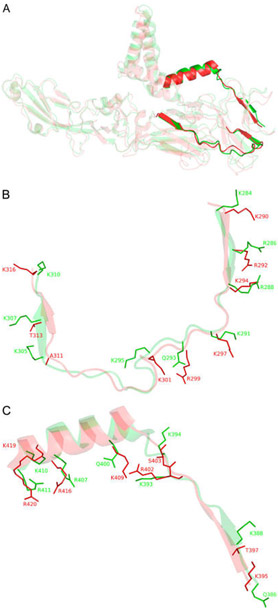
Figure 1.
(A) Full length structural alignment of ZIKV E (red, PDB entry 51RE) and DENV2 E (green, PDB entry 3J27). Bold structures are putative GAG-binding sites within the rest of the envelope proteins represented in more transparent structures. (B) Superimposition of putative GAG-binding site I of ZIKV E (K290–K316) and DENV2 E (K284–K310). Surface residues that may contribute to GAG binding are illustrated as sticks. (C) Putative GAG-binding site II of ZIKV E (R395–R420) and DENV2 E (Q386–R411) (see Figure S1 for a sequence alignment).