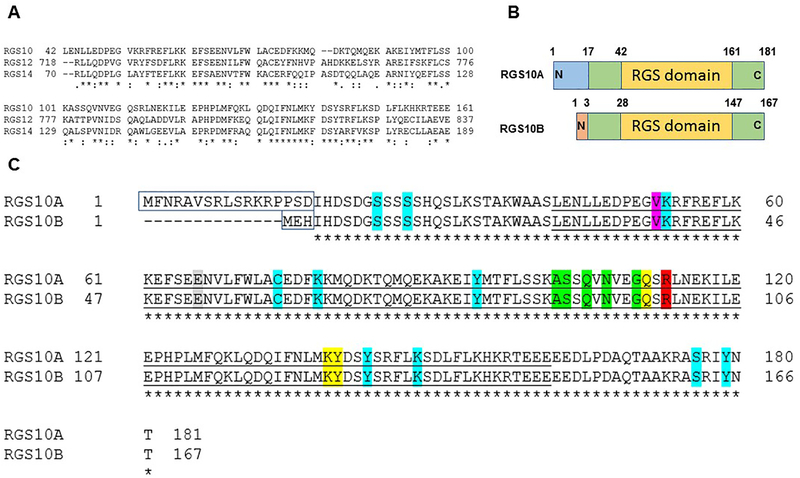
Figure 3: RGS10 isoforms: structure and analysis.
(A) Clustal Omega as a multiple sequence alignment program was used to align the RGS domain sequences within the D/R12 subfamily of human RGS proteins. (B) Schematic of the RGS10 isoform structures. (C) Alignment of human RGS10 isoform sequences using Clustal Omega program. Unfilled boxed amino acids represent N-terminal splice variants. Conserved catalytic RGS domains are underlined. The bright green highlighted residues, including (A102(88), S103(89), Q105(91), N107(93), G110(96)) are critical residues of RGS10 for the interaction with Gαi3 according to RGS10:Gαi3-AlF4− crystal structure (Protein Data Bank: 2IHB). The yellow-highlighted residues are putative disruptor residues, including Q111(97), K139(125), and Y140(126) that are predicted to impair the interaction with the Gα helical domain. The red-highlighted residue R113(99) is a conserved arginine residue within the RGS domain that does not contact the Gαi3 switch III region. The pink highlighted residue is a putative SNP that has been linked to schizophrenia. The gray-highlighted residue is the GAP dead mutation site based on the characterized GAP-dead mutation site in the RGS domain of RGS12 protein. Residue positions highlighted with turquoise are reported human RGS10 PTMs sites, including phosphorylation residues (S24(10), S27(13), Y94(80), Y143(129), S176(162), Y179(165)), ubiquitination residues (K53(39), K148(134)), palmitoylation C74(60) residue, and acetylation K78(64) residue.