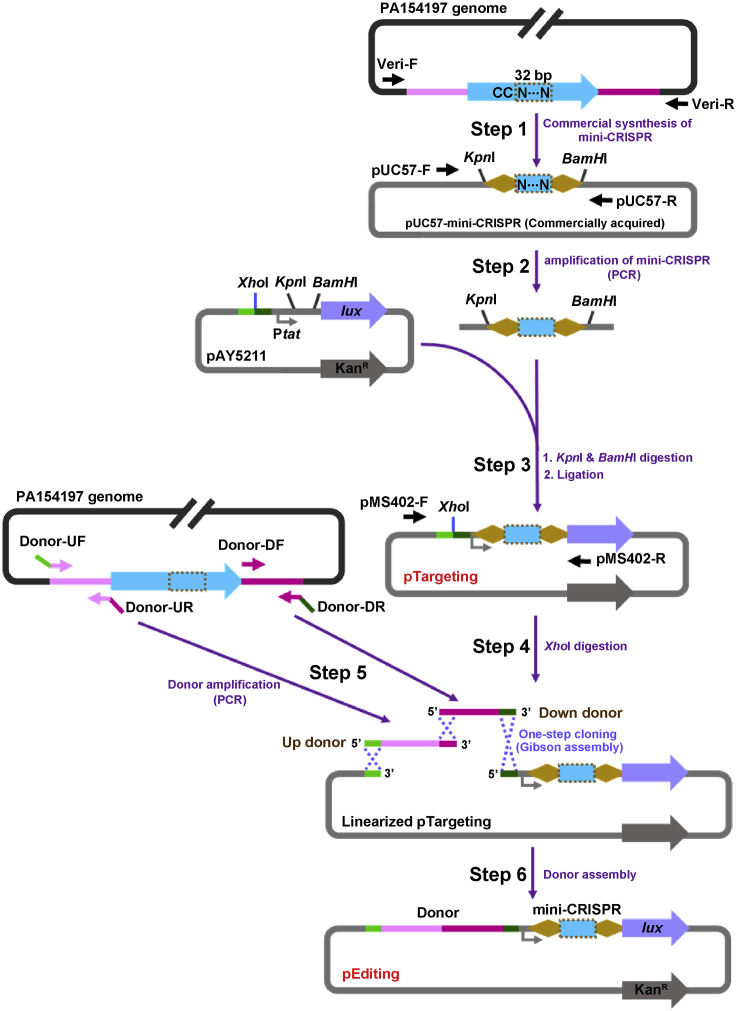
Figure 2.
Schematic Diagram of Constructing pTargeting and pEditing (Use Gene Deletion as an Example)
Step 1: An 88-bp mini-CRISPR which encompasses a 32-bp CC-preceding sequence (Dashed Box) within the target gene flanked by two 28-bp repeat sequences (Orange diamond) is submitted for commercial synthesis. Mini-CRISPR flanked by restriction sites KpnI and BamHI is supplied in the plasmid pUC57-mini-CRISPR.
Step 2: The mini-CRISPR is amplified by PCR using primers pUC57-F/R.
Step 3: After digestion with KpnI and BamHI, the purified mini-CRISPR is inserted into the plasmid pAY5211 which is treated with the same restriction enzymes, generating the targeting plasmid pTargeting. pTargeting is verified by sequencing using primers pMS-402-F/R.
Step 4: pTargeting is linearized by enzyme digestion with XhoI.
Step 5: 500-bp Up donor (Pink), containing a 15-bp 5′-end homology (Green) with the 3′ end of the linearized pTargeting and a 15-bp 3′-end homology (Magenta) with the Down donor, is amplified by PCR using primers Donor-UF/UR. 500-bp Down donor (Magenta), containing a 15-bp 5′-end homology with the 3′-end of the Up donor and a 15-bp 3′-end homology (Dark green) with the 5′ end of the linearized pTargeting, is amplified by PCR using primers Donor-DF/DR.
Step 6: Up Donor and Down donor are assembled into the linearized pTtargeting by Gibson assembly, generating the editing plasmid pEditing. pEditing is verified by sequencing using primers pMS-402-F/R. Detailed donor design for various types of genetic editing are presented in Figure 3.