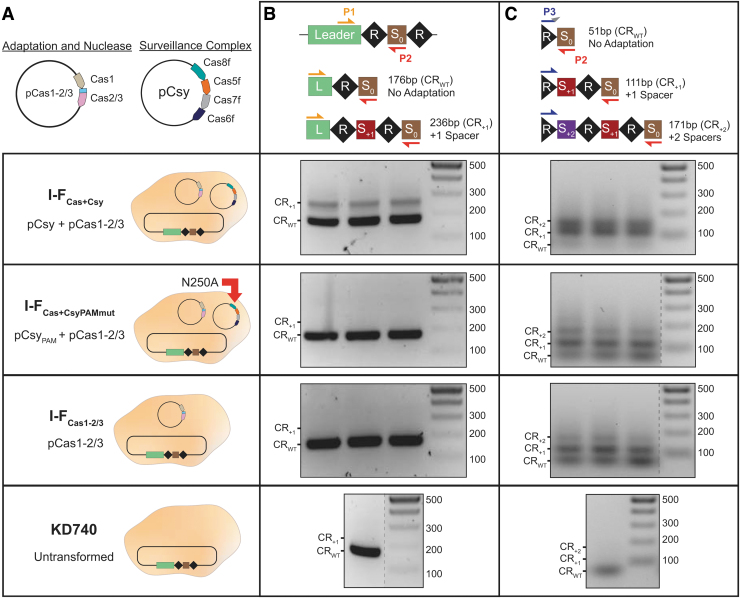
FIG. 1.
Cas1–2/3 proteins are necessary and sufficient for new spacer acquisition, but Csy accelerates adaptation. (A) Schematics of the Cas1–2/3 and Csy subunit (Cas8f, Cas5f, Cas7f, and Cas6f) expression vectors are shown above schematics for each of the conditions tested. Escherichia coli KD740 cells are shown as tan ovals. (B) PCR amplification with “internal” (P1 and P2) primers to detect adaptation events. (S0 is initial spacer that targets phage λ.) Expected sizes of PCR products for CRISPR with acquired spacers (CR+1 and CR+2) or no acquisition (CRWT) are labeled. Adaptation events detected after PCR with primers P1 and P2. Each lane represents a biological replicate. (C) PCR products from (B) were enriched for higher molecular weight species, and then used as templates for a subsequent round of CAPTURE PCR with primers P3 and P2. Products corresponding to expanded arrays are visible in all biological replicates for I-FCas+Csy, I-FCas+CsyPAMmut, and I-FCas1–2/3. Cas, CRISPR-associated protein; PAM, protospacer adjacent motif; PCR, polymerase chain reaction.