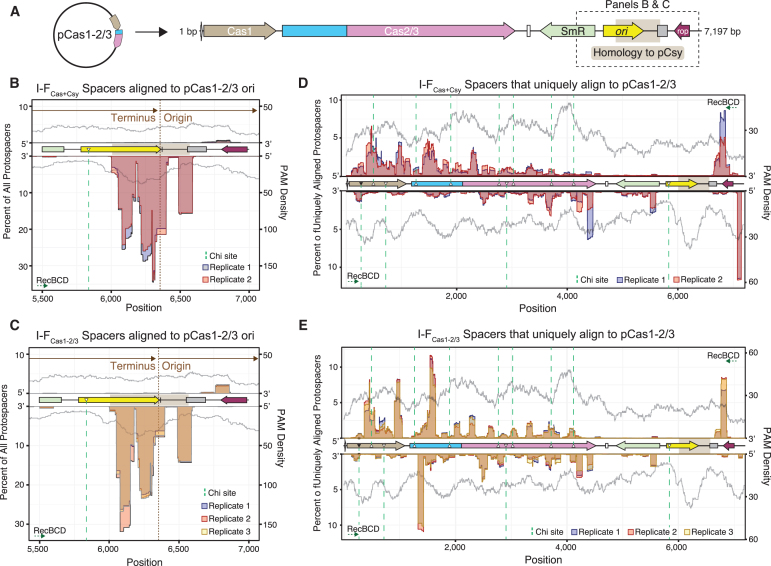
FIG. 3.
Protospacer hotspots are reproducible across multiple biological replicates. (A) Linearized genetic map of Cas1–2/3 expression plasmid. Regions shown in (B, C) are marked with dotted outline. (B, C) Positions and quantities of all protospacers that align to ori of pCas1–2/3, including spacers that produce alignments to additional sequences (e.g., pCsy). Protospacer locations are mapped using a 50 nt sliding window. Data from each biological replicate are represented as a different color. PAM densities (gray) mapped with a 150 nt sliding window. Direction and start/end positions of plasmid replication shown in brown. Consensus Chi sites are shown as triangles filled in with black, and Chi sites with a single base mismatch are shown as triangles with a white fill. Dotted green lines indicate the boundaries of Chi sites, and RecBCD directions of cleavage are shown for each strand of DNA. (D, E) Positions and quantities of protospacers that produce a single unique alignment to pCas1–2/3 are shown as in (B, C).