Fig. 5.
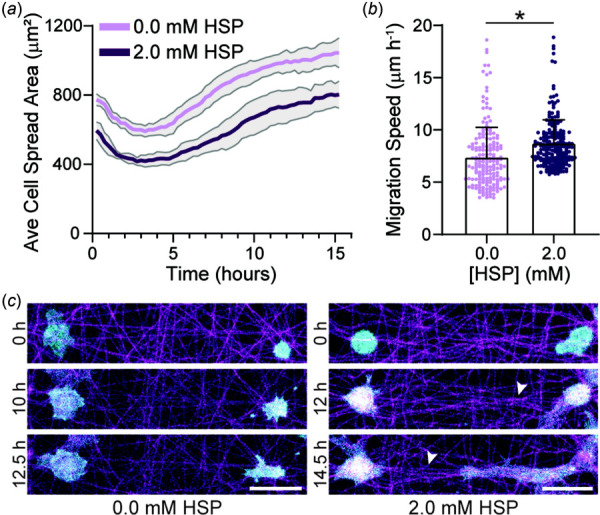
EC spreading and migration dynamics on crimped DexVS matrices. (a) Average cell spread area over 16 h after seeding on DexVS matrices functionalized with various HSP concentrations (n = 6 fields of view). (b) Migration speed as a function of HSP concentration (n ≥ 197 cells). (c) Representative time-lapse images of lifeAct-GFP expressing ECs on DexVS matrices functionalized with 0.0 and 2.0 mM HSP. Arrows indicate matrix alignment between interacting cells. F-actin (cyan), DexVS fibers (magenta). Scale bars: 50 μm. All data presented as mean ± standard deviation; *p < 0.05.
