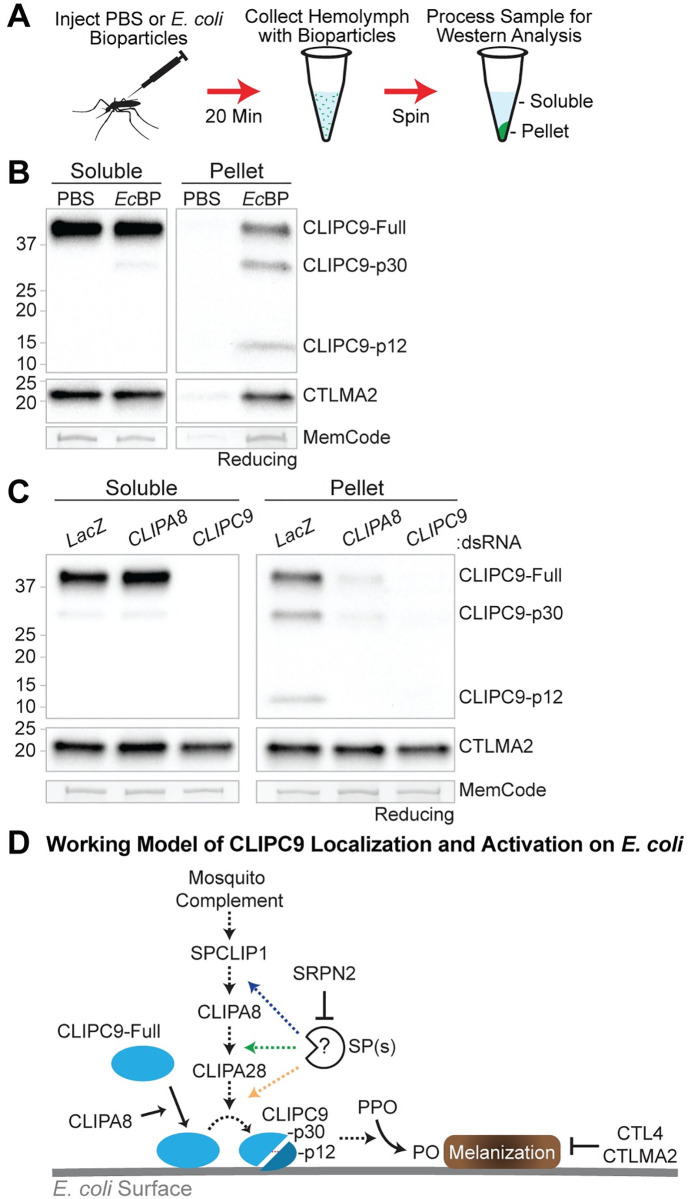
Fig 10. CLIPA8 regulates the localization and cleavage of CLIPC9.
(A) The E. coli bioparticles (EcBP) surface extraction model was utilized to assess the localization of CLIPC9 to bacterial surfaces 20 min post-challenge. PBS injection served as a negative control. (B) Reducing western analyses of CLIPC9 on soluble and pellet fractions following PBS or EcBP injection. Blots were probed against CTLMA2 and MemCode stained to confirm sample reduction and protein loading, respectively. Western image is representative of three independent biological replicates. (C) Reducing western analyses of CLIPC9 in soluble and pellet fractions obtained from dsLacZ-, dsCLIPA8-, and dsCLIPC9-treated mosquitoes following EcBP injection. CTLMA2 staining was utilized as a localized immune protein loading control and to verify sample reduction. MemCode staining was used to confirm equal protein loading. Image is representative of experiments from three independent biological replicates. (D) Working model illustrating the CLIPA8-dependent recruitment of CLIPC9 to E. coli surfaces, its activation downstream of the mosquito complement-dependent CLIP-SPH pathway, and its negative regulation by SRPN2. The CTL4/CTLMA2 heterodimer negatively regulates E. coli-induced PO activity64, however its relationships with mosquito complement, the CLIP-SPH pathway, and CLIPC9 are unknown. Dashed arrows indicate uncharacterized enzymatic steps.