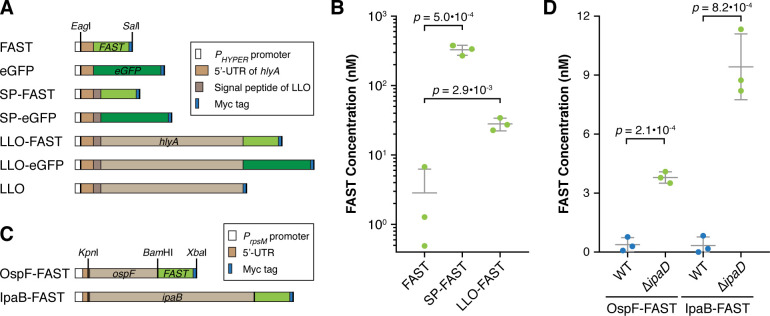
Fig 1. FAST-tagged proteins retain fluorescent properties after secretion into bacterial culture media.
(A) Diagram of constructs in the pAD vector for constitutive expression in Lm. (B) Lm strains expressing FAST-tagged proteins were cultured in LSM, then fluorescence intensities were measured on the filtered supernatants of each culture in presence of 5 μM HBR-3,5DM. Concentrations of FAST-labelled proteins were calculated by reference to a standard curve of purified FAST in LSM. Residual fluorescence measured for the strain producing non-secreted FAST represents bacterial lysis. (C) Diagram of constructs in the pSU2.1 vector for expression in Sf. (D) WT or ΔipaD Sf strains expressing FAST-tagged OspF or IpaB were cultured in M9 medium, then fluorescence intensities were measured on the filtered supernatants of each culture in presence of 5 μM HBR-3,5DM. Concentrations of FAST-labelled proteins were calculated by reference to a standard curve of purified FAST in M9 medium. Whereas ΔipaD strains secrete proteins constitutively, T3SS secretion is not activated in WT strains, thus the fluorescent signals measured for these strains (blue dots) reflect bacterial lysis and/or leakage of the T3SS. (B, D) Normalized values, means and standard deviations from three independent experiments were plotted. p-values represent the results of two-tailed Student’s t-tests with equal variance assumption. Source data are provided in S3 Table.