Fig 4. HCProWD is defective in recruiting VCS to PGs.
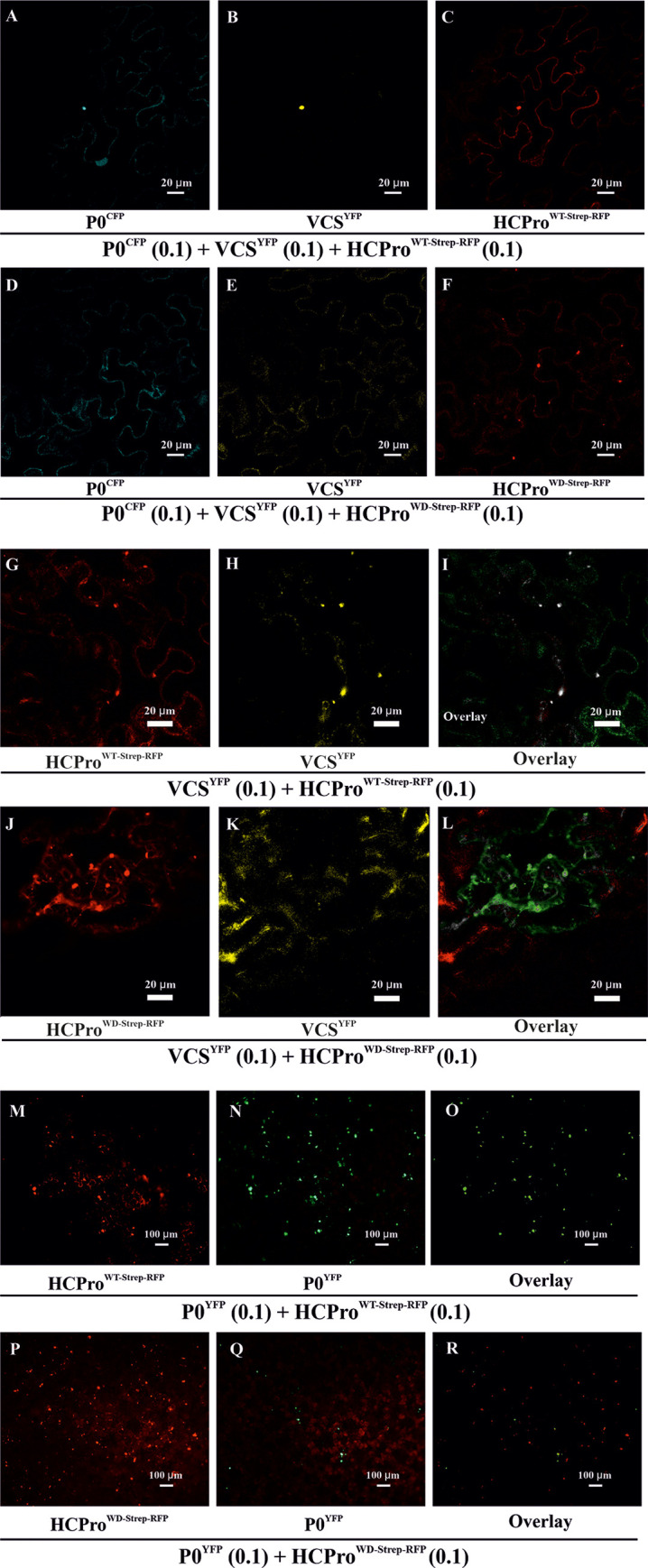
(A-F) Localization of Strep-RFP-tagged -HCPro, YFP -tagged-VCS and CFP tagged -P0 in N. benthamiana epidermal cells. Agrobacterium carrying either HCProWT-Strep-RFP or HCProWD-Strep-RFP was infiltrated at OD600 = 0.1, while Agrobacteria harboring VCS-AYFP, VCS-BYFP and VCS-CYFP were each infiltrated at OD600 = 0.1 (together denoted as VCSYFP). Agrobacterium harboring P0CFP, infiltrated at OD600 = 0.1, was expressed as another marker for PGs. Samples were analyzed at 3 dpi by confocal laser scanning microscopy in sequential scanning mode. (A) P0CFP, (B) VCSYFP, (C) HCProWT-Strep-RFP, (D) P0CFP, (E) VCS-YFP and (F) HCProWD-Strep-RFP. (G-L) Localization of HCProWT/WD-Strep-RFP and VCSYFP and their overlays in N. benthamiana epidermal cells. Experimental details are otherwise the same as in Fig 4A–4F, except for the absence of P0CFP. (G) HCProWT-Strep-RFP, (H) VCS-YFP and (I) Overlay of HCProWT-Strep-RFP and VCSYFP. (J) HCProWD-Strep-RFP, (K) VCSYFP and (L) Overlay of HCProWD-Strep-RFP and VCSYFP. Degree of co-localization between HCProWT-Strep-RFP / HCProWD-Strep-RFP and VCSYFP is calculated from representative images and presented in S6 Fig. (M-R) Epifluorescence microscopy showing the distribution of HCProWT/WD-Strep-RFP and P0YFP within a representative area of examination. HCPro and P0YFP, which both typically co-localize in PGs, were co-infiltrated to visualize the PGs. The experiment was conducted similarly as Fig 3E and 3F. The only exception was that HCProWT/WD-Strep-RFP was used instead of HCProWT/WD. Images were acquired at 3 dpi with a 10X objective. HCProWT/WD-Strep-RFP was visualized under an RFP filter (M, P respectively). P0YFP from the corresponding sample sets was visualized under the FITC filter (N, Q respectively), and their overlays are presented in (O, R), respectively.
