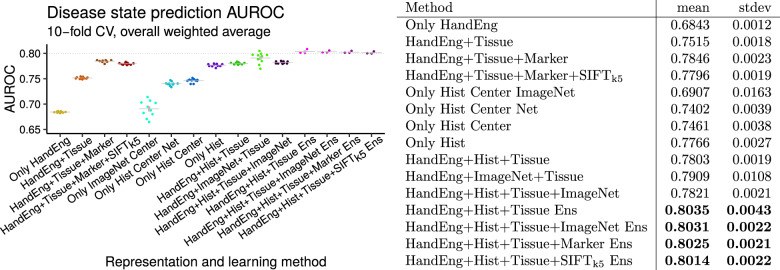
Fig. 9. Disease state prediction performance for machine learning methods..
For deep learning we use a ResNet-50. For shallow learning we use a Random Forest. We train a Random Forest on deep features (and other features), to combine deep and shallow learning (Fig. 3C top). Error bars indicate standard error of the mean. Points indicate replicates. Gray lines indicate means. Performance increases markedly when including tissue type covariate for learning (even though tissue type is missing for some patients), when using deep learning to integrate information throughout entire image rather than only the center crop, and when using an ensemble of classifiers. Performance exceeds AUROC of 0.8 (at right). We conclude method xii (“HandEng + Hist + Tissue Ens”) is the best we tested for disease state prediction, because no other method performs significantly better and no other simpler method performs similarly. Methods are, from left to right, (i) Random Forest with 2412 hand-engineered features alone for 512 × 512 px scaled and cropped center patch, (ii) Random Forest with tissue covariates, (iii) Random Forest with tissue and marker covariates, (iv) method iii additionally with SIFTk5 features for Random Forest, (v) only natural-image-trained ResNet-50 at same scale as method i with center 224 × 224 px center patch and prediction from a Random Forest trained on 2048 features from the ResNet-50 (Fig. 3), (vi) histopathology-trained ResNet-50 at same scale as method i with center 224 × 224 px center patch and prediction from top three neurons (Fig. 3B top), (vii) histopathology-trained ResNet-50 with Random Forest trained on 100 features from 224 × 224 px center patch per method vi, (viii) histopathology-trained ResNet-50 features at 21 locations throughout image summed and Random Forest learning on this 100-dimensional set representation with 2412 hand-engineered features, (ix) method viii with tissue covariates for histopathology-trained ResNet-50 and 2412 hand-engineered features for Random Forest learning (i.e., Fig. 3C sans marker information), (x) method ix with an only natural-image-trained ResNet-50 instead of a histopathology-trained ResNet-50 for Random Forest learning, (xi) method ix with both an only natural-image-trained ResNet-50 and a histopathology-trained ResNet-50 for Random Forest learning, (xii) method ix with an ensemble of three Random Forest classifiers such that each classifier considers an independent histopathology-trained ResNet-50 feature vector in addition to 2412 hand-engineered features and tissue covariate, (xiii) method xii where each Random Forest classifier in ensemble additionally considers only natural-image-trained ResNet-50 features, (xiv) method xii where each Random Forest classifier in ensemble additionally considers the marker mention covariate (i.e., this is an ensemble of three classifiers where Fig. 3C is one of the three classifiers), (xv) method xii where each Random Forest in ensemble additionally considers SIFTk5 features for learning.