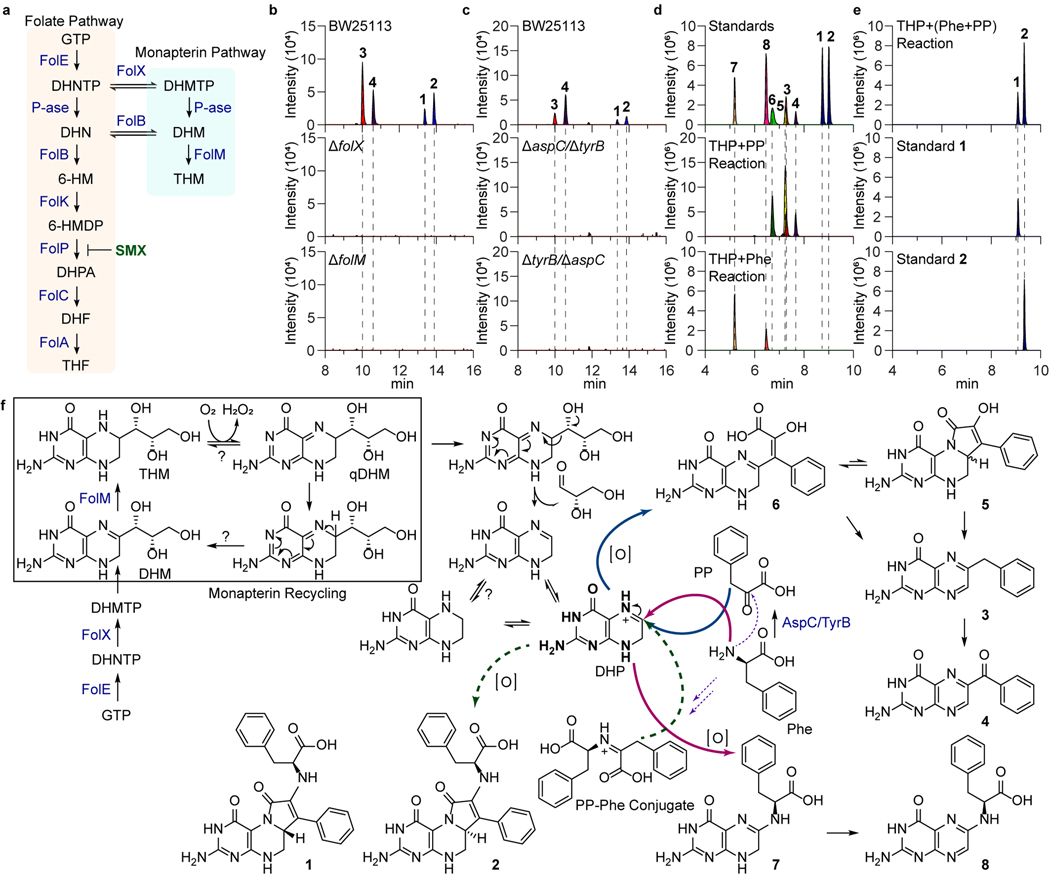
Fig. 2. Genetic and biomimetic synthesis support of the colipterin pathway.
a, Schematic representation of the folate and monapterin pathway. Chemical structures and enzyme annotations of these pathway are given in Extended Data Fig. 4a. b, Abolishment of major colipterins 1-4 in monapterin biosynthetic mutants, ΔfolX and ΔfolM. c, Abolishment of 1-4 in two distinct double-mutants of aspC and tyrB in E. coli BW25113. d, Production of colipterins 3-8 via in vitro biomimetic synthesis. Tetrahydropterin (THP) was used as a dihydropterin (DHP) substrate, which reacts with phenylpyruvate (PP) to yield 3-6 (middle) and L-phenylalanine (L-Phe) to yield 7 and 8 (bottom), respectively. e, Pre-incubated phenylpyruvate (PP) and L-phenylalanine (L-Phe) reactions with THP yielded 1 and 2. f, Proposed colipterin pathway in E. coli. Proposed monapterin recycling is boxed. Enzyme annotations (blue colored) are as follows: FolE, GTP cyclohydrolase I; FolX, 7,8-dihydroneopterin triphosphate 2ʹ-epimerase; and FolM, 7,8-dihydromonapterin reductase. Abbreviations are as follows: GTP, guanosine triphosphate; DHNTP, 7,8-dihydroneopterin triphosphate; DHMTP, 7,8-dihydromonapterin triphosphate; DHM, 7,8-dihydromonapterin; THM, Tetrahydromonapterin; and qDHM, quinoid dihydromonapterin. Solid blue and red arrows indicate coupling of PP and L-Phe to DHP, respectively. Dashed arrows represent the formation of PP-Phe conjugate (purple) and coupling of this conjugate to DHP (green), respectively.