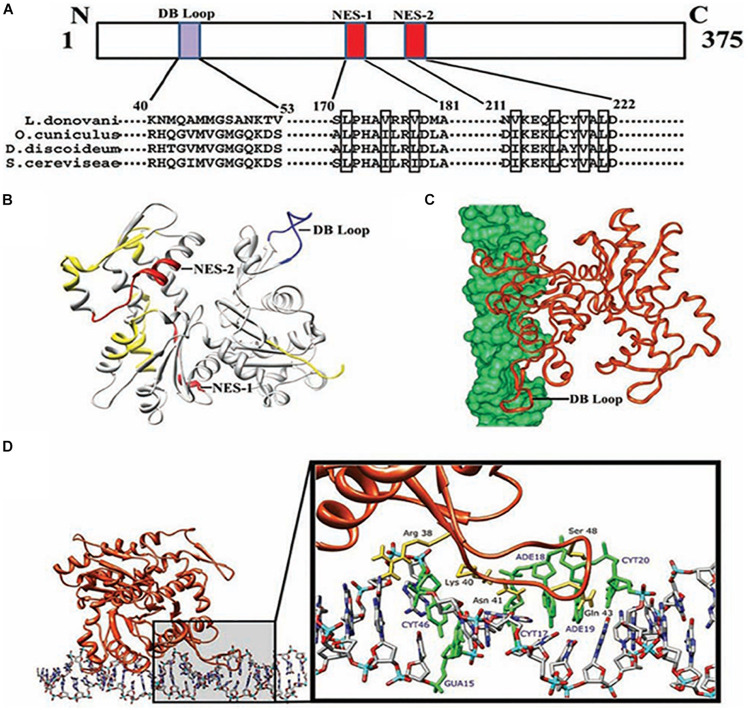
FIGURE 5.
Computational docking of average simulated model of LdAct with DNA showing the interaction of the diverged DB-loop of LdAct with the major groove of DNA. (A) Sequence alignment of LdAct with other actins showing the presence of nuclear export signals (NES-1 and NES-2) in the LdAct aa sequence and the diverged DB-loop predicted to be involved in the DNA binding, by DP-Bind server. (B) Energy minimized average simulated model of LdAct showing positions of NES-1, NES-2 (red) and the diverged stretches of amino acid sequences (yellow) including the sequence that fall in DB loop (blue). (C) Docking of LdAct (orange) with DNA (green) using HADDOCK protocols. (D) Amino acid residues of the DB loop of LdAct (yellow) showing hydrogen bonding with the nucleotides (green) of DNA. DB, DNase I binding; NES, nuclear export signal (This was originally published in Nucleic Acids Research, Kapoor et al., 2010© Oxford University Press).