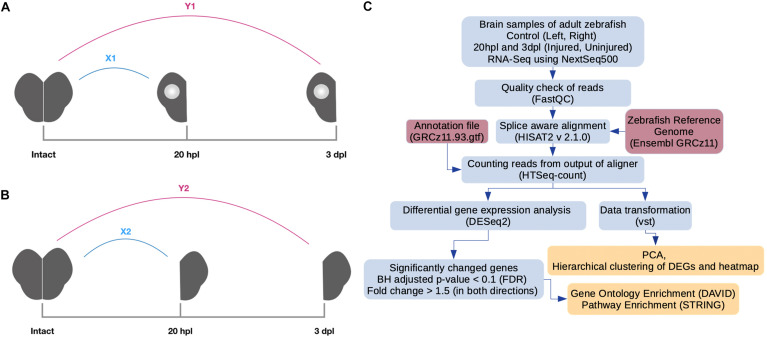
FIGURE 1.
Pipeline for RNA preparation and analysis of RNA sequencing (RNA-seq) data. (A) Preparation of RNA samples from the lesioned hemispheres at 20 hpl or 3 dpl. X1: comparison of the lesioned hemisphere at 20 hpl with the intact brain; Y1: comparison of the lesioned hemisphere at 3 dpl with the intact brain. (B) Preparation of RNA samples from the unlesioned hemispheres at 20 hpl or 3 dpl. X2: comparison of the unlesioned hemisphere at 20 hpl with the intact brain; Y2: comparison of the unlesioned hemisphere at 3 dpl with the intact brain. (C) RNA-seq reads were generated using Illumina NextSeq 500. The quality of reads was checked by the FastQC tool. Next, alignment and counting of reads were performed by using a zebrafish reference genome (Ensembl, GRCz11) and the annotation file (Ensembl, GRCz11.93) by HISAT2 and HTSeq-count tools, respectively. Counts were transformed with variance stabilizing transformation (vst) to perform principal component analysis and hierarchical clustering. To select genes as differentially expressed, we used the DESeq2 package with the following criteria: fold change > 1.5 (in both directions) and Benjamini–Hochberg adjusted p-value (FDR) < 0.1. Gene Ontology and pathway enrichment analyses were performed by using DAVID and the STRING tool of Cytoscape, respectively. hpl, hours post-lesion; dpl, days post-lesion.