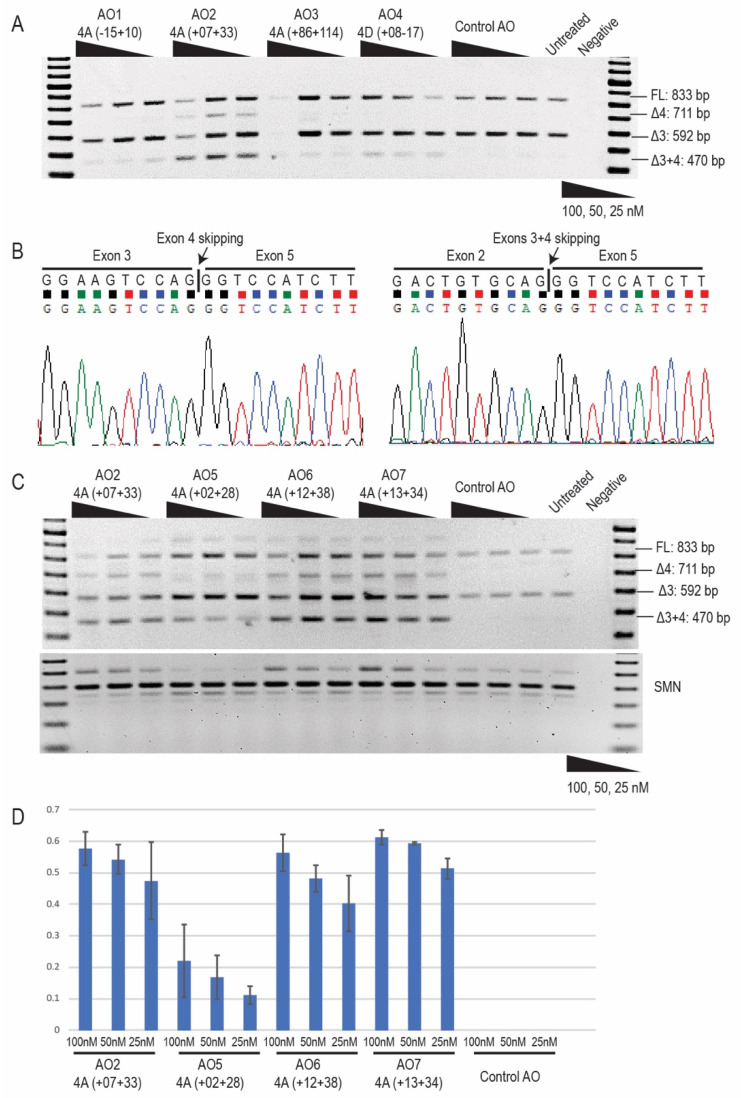
Figure 2.
PRKN transcript analysis following 2′-O-methyl PS AO transfection. (A) RT-PCR analysis of PRKN transcripts from Parkinson’s patient-derived fibroblasts with genomic deletion of exon 3, following transfection with PRKN exon 4 targeting 2′OMe PS AOs at concentrations of 100, 50, and 25 nM. The control AO that does not anneal to any transcript was used as a negative control. A 100 bp DNA ladder was used to indicate amplicon sizes, and an RT-PCR no-template negative control was loaded in the last lane. (B) Sanger sequencing identified skipping of exon 4 on the allele carrying the missense mutation and the absence of exons 3 + 4 on the allele with PRKN exon 3 genomic deletion. (C) RT-PCR analysis of PRKN transcripts after the transfection of “microwalking” AOs. The original AO sequence, AO2, was included for comparison of exon skipping efficiencies. SMN transcript was used as a house-keeping gene and normalization of PRKN transcripts. (D) Densitometric analysis of the Δ3 + 4 band showed a dose-dependent response to all of the AOs and indicated the AO sequence that induced the highest level of exon skipping. FL; full-length.