Table 6.
Motifs found in the promoters of selected differentially expressed genes (DEGs) exclusively found in DH372 in the Multiple Expectation maximization for Motif Elicitation (MEME) analysis: the top 5 exclusive motifs identified by MEME in promoters presented here with their E-value. The Gene Ontology for Motifs (GOMO) analysis provides information regarding the novelty of the motif and the probability that this motif is involved in certain cellular functions.
| MEME Identified Motif Logo | Consensus Motif in Common Responding Genes | E-Value | Number of GO Terms Identified by GOMO | Role of Motif Identify by GOMO |
|---|---|---|---|---|
1-
|
TTWTTTWTYBBYWWSTTTTTBTSTC | 1.1 × 103 | 05 |
MF transcription factor activity CC nucleus CC plasma membrane MF protein binding BP regulation of transcription, |
2-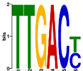
|
TTGACY | 3.6 × 104 | 01 | MF transcription factor activity (WRKY TFs) |
3-
|
TKTADATKTRTSATGRBTGCT | 6.9 × 104 | 0 | |
4-
|
CTCCATGATTG | 6.1 × 104 | 0 | |
5-
|
AARCAAVGVAGCMAC | 1.2 × 105 | 0 |
The E-value is an estimate of the expected number of motifs with the given log likelihood ratio (or higher) and with the same width and site count that one would find in a similarly sized set of random sequences (sequences where each position is independent and letters are chosen according to the background letter frequencies). The consensus sequences are presented in the International Union of Pure and Applied Chemistry (IUPAC) nomenclature and constructed from each column in a motif’s frequency matrix using the “50% rule”.
