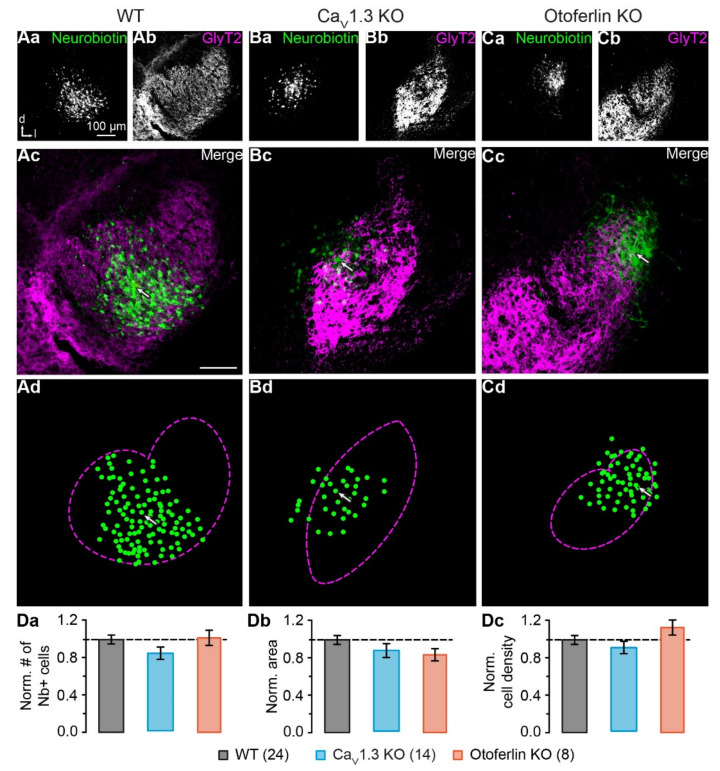
Figure 3.
Reconstruction of LSO astrocyte networks. (A–C), tracer-coupled networks of the WT, CaV1.3 KO, and otoferlin KO mice. The tracer neurobiotin diffused from the patch-clamped astrocyte into coupled cells (Aa–Ac,Ba–Bc,Ca–Cc). Immunohistochemical labeling for glycine transporter (GlyT) 2 highlighted the LSO (Ab,Bb,Cb) and allowed the localization of the network within the nucleus (Ac,Bb,Cc). Cells with fluorescence intensity of at least 1.75-fold background intensity were transferred to a schematized representation and are displayed by green dots (Ad,Bb,Cd). The dotted magenta lines indicate the LSO border as derived from GlyT2 labeling (Ab,Bb,Cb). The arrows in (Ac,Ad,Bc,Bd,Cc,Cd) mark the patched cell. (D), network properties. Values were normalized to the WT data, indicated with the dashed line. There were no differences between the number of coupled cells (Da), network area (Db), or density of coupled cells (Dc). The WT data (Aa–Ad) were part of [9]. Panel (Aa–Ac) was reused from that publication. (Da–Dc) show mean ± SD. Number of slices is given in parentheses.