Figure 1.
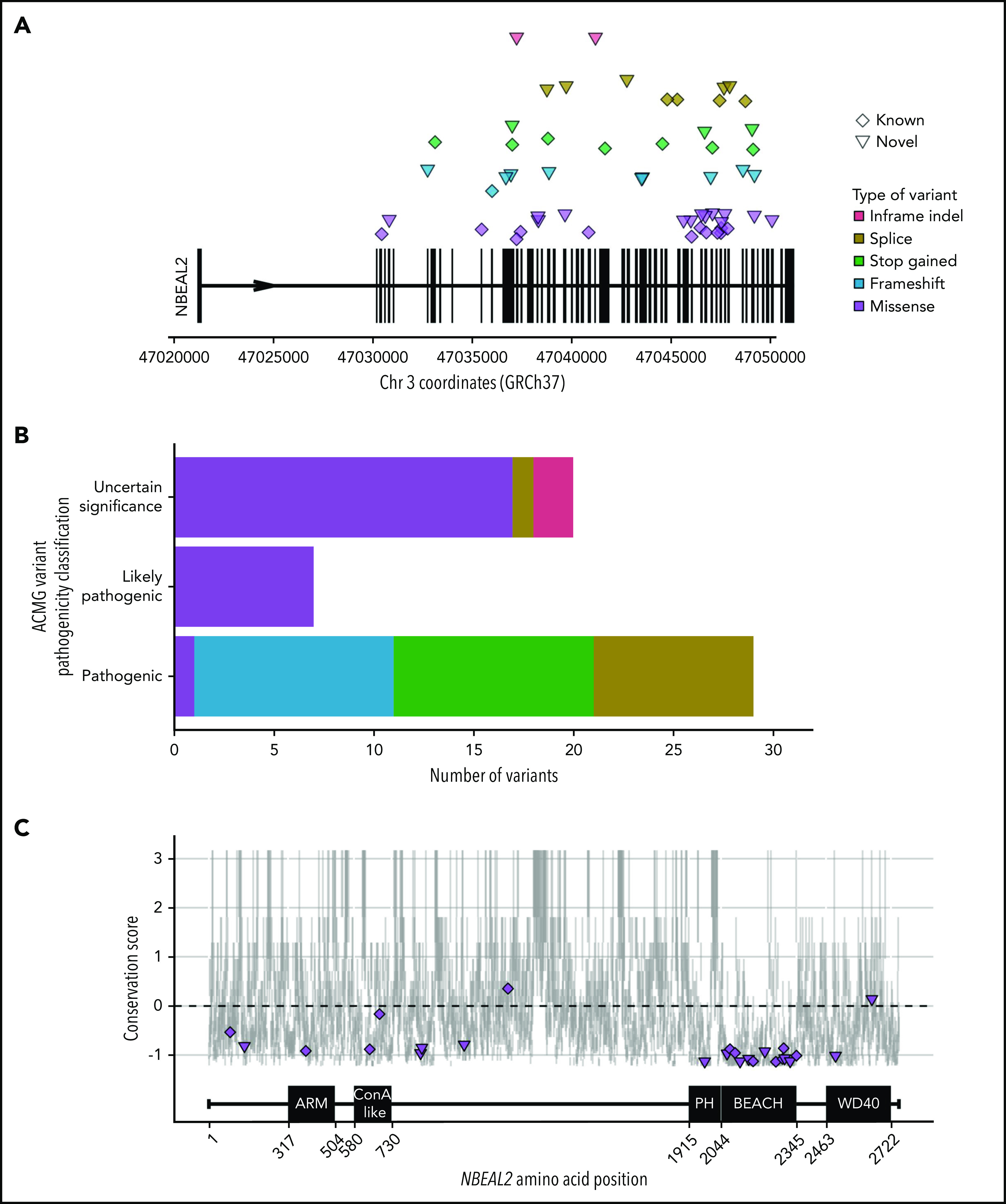
NBEAL2 variants identified in patients with GPS participating in this study. (A) Position of the 56 unique variants relative to NBEAL2 on chromosome 3 (Chr 3) using Genome Reference Consortium human genome build 37 (GRCh37). Vertical black bars represent exons. The color and shape used to represent variants in the legend apply throughout the figure. Inframe indel refers to variants which are inframe small insertions and/or deletions ≤50 base pairs. (B) The frequency of unique variants classified by pathogenicity, according to the American College of Medical Genetics and Genomics (ACMG) guidelines.22 (C) The location of missense variants compared with the amino acid sequence of Nbeal2 and known functional domains (on the x-axis). The y-axis position of the variant is the estimated evolutionary conservation score of the affected residue calculated by using ConSurf.37 A positive score corresponds to lower conservation, and a negative score represents higher conservation. Vertical gray lines represent the range of conservation scores of the 50% confidence interval: the bottom and top of the bar are the 25th and 75th percentile, respectively, of the inferred evolutionary rate distribution.
