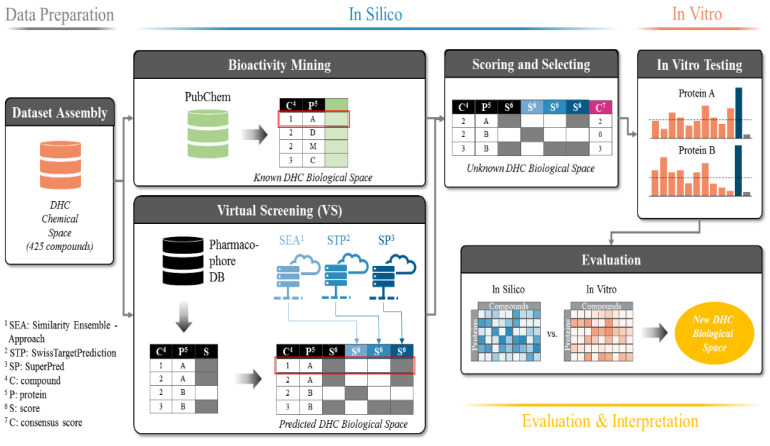
Figure 1.
Workflow of the dihydrochalcone (DHC) target prediction campaign. The dataset is assembled (DHC chemical space) and used to retrieve corresponding bioactivity data from PubChem (known DHC biological space) and as input to inverse VS. First, the DHC chemical space is mapped onto the Pharmacophore DB (database) and the resulting matrix extended by the predictions of three individual target prediction servers: Similarity Ensemble Approach (SEA), SwissTargetPrediction (STP), and SuperPred (SP), resulting in the predicted DHC biological space. Activities already known from PubChem (the known DHC biological space) are then removed from the predicted DHC biological space, and the reduced matrix is scored according to the consensus predictions of the ligand-target interactions (unknown DHC biological space). Protein targets of the unknown DHC biological space are selected according to their consensus score (CS) and chosen for in vitro biological evaluation.