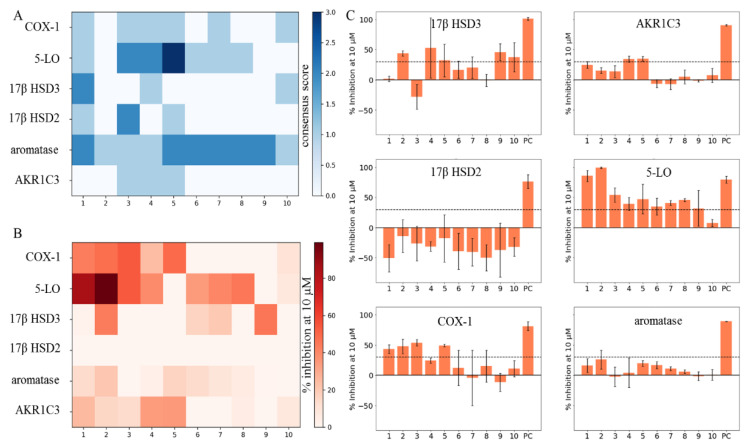
Figure 4.
Comparison of an unknown DHC biological space (COX-1, 5-LO, 17β HSD3, 17β HSD2, aromatase, and AKR1C3) and actual in vitro test results of compounds 1–10. (A) CSs of compounds 1–10 on all of the six targets of the unknown DHC biological space plotted as a heatmap. (B) Means (n = 3) of the percent inhibition at 10 µM (0–100%) of compounds 1–10 on all of the six targets of the unknown DHC biological space plotted as a heatmap. Observations with mean inhibition values smaller than 30% or relative standard deviations greater than 20% were regarded as inactive. (C) Bar charts of the six targets of the unknown DHC biological space showing compounds 1–10 with the respective means (n = 3) of the percent inhibition at 10 µM (0–100%) and the standard deviation. A cut-off of 30% inhibition at 10 µM was chosen (black dashed line), for separating the active from inactive observations. Dimethylsulfoxide (DMSO) was used to measure the baseline enzyme activities on which the samples were normalized (not shown), and positive controls (PC) were used as indicated in the Materials and Methods and in Supplementary Information S-5.