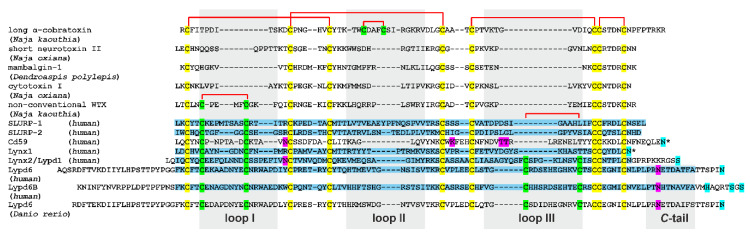
Figure 1.
Amino acid sequence alignment of three-finger proteins (TFPs). Representative snake toxins from various families, several human TFPs, and fish Lypd6 are shown. The signal peptides are removed. Invariant Cys residues are shown in yellow. Cys residues forming additional disulfide bonds in the loops I, II, and III are shown in green. Disulfide bonds are shown by red brackets. Predicted sites for attachment of the GPI-anchor and glycosylation are shown by cyan and magenta, respectively. Several possible GPI-anchor sites were predicted for human Lypd6b. Fragments of the proteins used for NMR studies in this and previous works [11] are highlighted in blue. The proteins, for which the presence of both the GPI-anchored and soluble form were reported, are marked by an asterisk. The loop regions and C-terminal regions used for calculation of mean RMSD and S2 values are highlighted by gray background. PDB codes: α-cobratoxin—2CTX, neurotoxin II—2MJ4, mambalgin-1—5DU1, cytotoxin I—5NPN, WTX—2MJ0, SLURP-1—6ZZE/6ZZF, SLURP-2—2N99, CD59—2J8B, Lynx1—2L03, Lynx2—6ZSS, Lypd6—6IB6, Lypd6B—6ZSO. The structure of Danio rerio Lypd6 is not available.