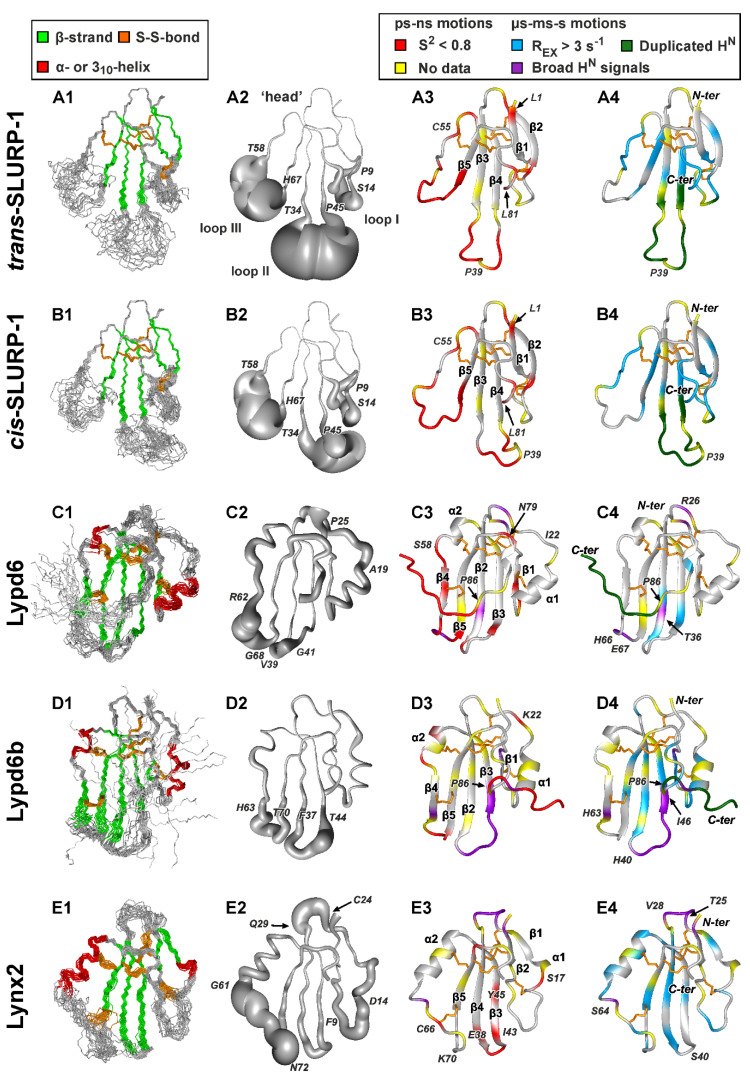
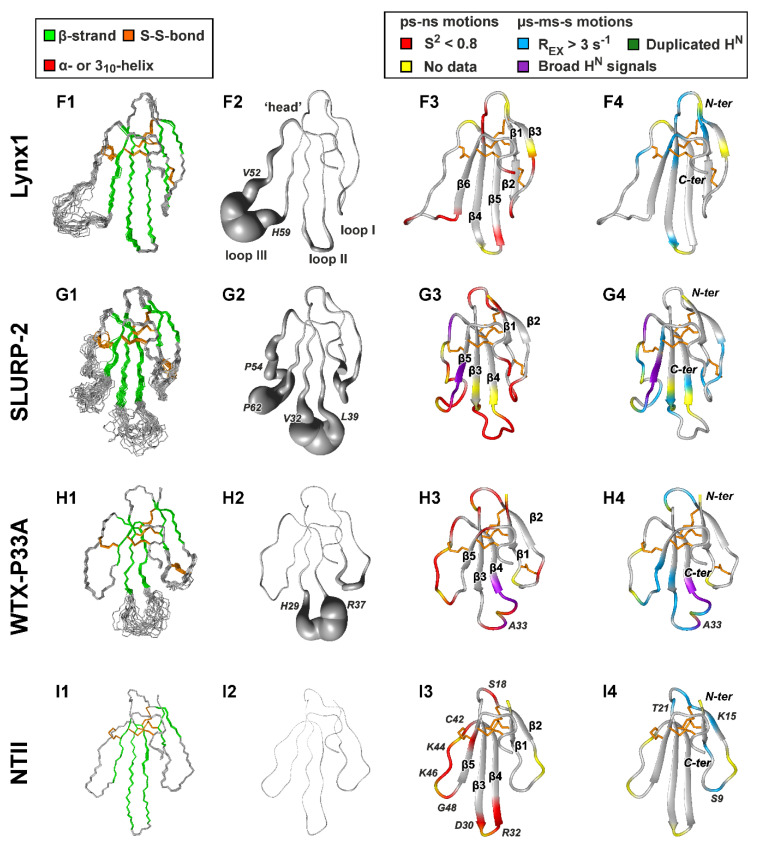
Figure 2.
NMR structures and dynamics of TFPs: (A) trans-Tyr39-Pro40 SLURP-1, (B) cis-Tyr39-Pro40 SLURP-1, (C) Lypd6, (D) Lypd6b, (E) Lynx2, (F) Lynx1, (G) SLURP-2, (H) WTX-P33A, (I) NTII. For each protein/isomer, four panels are shown. (1) Set of the 20 best CYANA structures. Protein backbone and disulfide bonds (gold) are shown. Secondary structure elements are color-coded: red—α- or 310-helix, green—β-sheet. (2) ‘Sausage’ representation of the mean structure with variable radius equal to the average displacement in the set of structures. In the Lypd6 and Lypd6b structures, the completely disordered C-terminal fragments (Leu85-Ala95) are omitted. (3–4). Ribbon representation of NMR structures with mapped data about ‘fast’ (ps–ns, 3) and ‘slow’ (µs-ms-s, 4) backbone dynamics. Regions with the high-amplitude ps–ns mobility (generalized order parameter S2 < 0.8) are in red. Sites of µs-ms-s conformational fluctuations are in blue (exchange contributions to R2 relaxation rates Rex > 3 s−1 at 800 MHz), violet (significant broadening of 1HN signals), and dark green (two protein conformers were observed). N-terminal Met0 residue, Pro residues, and the residues for which dynamics data are unavailable due to spectral overlap are highlighted by yellow. The residues from the disordered C-terminal fragments of Lypd6 and Lypd6b, which did not satisfactorily fit into any model of intramolecular mobility, are shown in red. Structural data for Lynx1, SLURP-2, WTX-P33A, and NTII were taken from previous publications [11,32,43,44]; other data are from the present work. PDB codes: WTX-P33A—2MJ0, SLURP-1—6ZZE/6ZZF, SLURP-2—2N99, Lynx1—2L03, Lynx2—6ZSS, Lypd6—6IB6, Lypd6B—6ZSO.