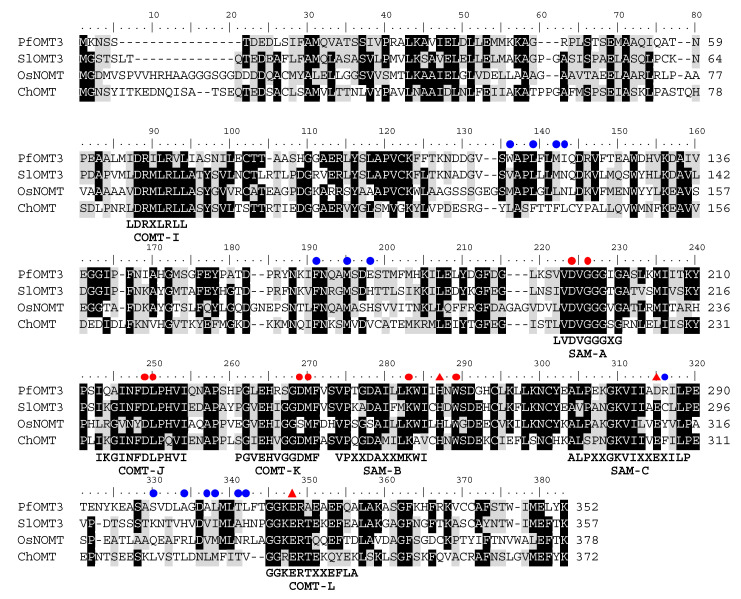
Figure 1.
Multiple alignment of Perilla frutescens O-methyltransferase3 (PfOMT3) amino acid sequence with other class II O-methyltransferases (OMTs). Amino acid sequences of Solanum lycopercicum OMT3 (SlOMT3), Oryza sativa naringenin OMT (OsNOMT) and chalcone OMT (ChOMT) from alfalfa were used in the alignment. Three S-adenosyl-L-methionine (SAM)-binding motifs (SAM-A, B and C) and four caffeic acid OMT (COMT) motifs (COMT-I, J, K and L) conserved in class II OMTs are indicated with their consensus sequences under the aligned sequences. Red triangles indicate the three catalytic residues of class II OMTs. Red and blue circles indicate SAM-binding residues and substrate-binding residues, respectively. Identical and similar residues are shaded in black and gray, respectively.