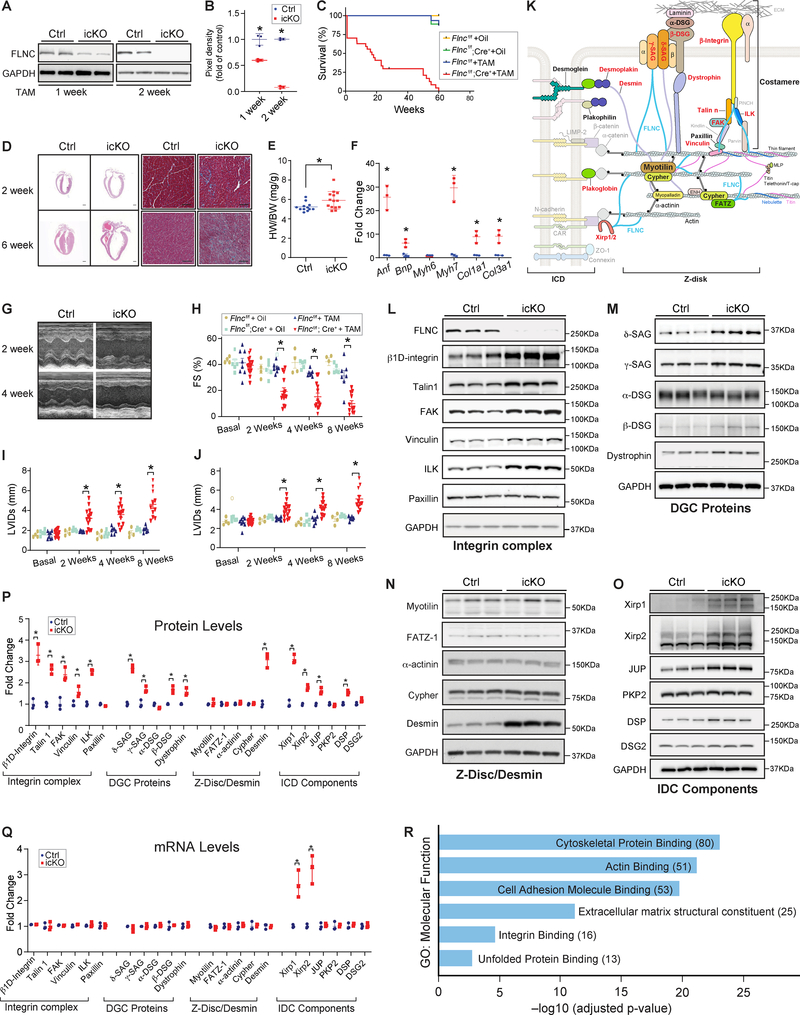
Figure. Loss of Filamin C is catastrophic for heart function.
(A-B) Representative western blot (A) and quantitation (B) showing efficient loss of Flnc in icKO mouse hearts 2 weeks post tamoxifen treatment (TAM) (40 mg/kg/day for 3 days). Flncf/f, Cre− littermates post TAM served as controls (Ctrl). n= 3 per group. (C) Kaplan-Meier survival curves of Flncf/f and Flncf/f; Cre+ mice injected with TAM or vehicle control. n= 16, 8, 26, and 9 respectively. (D) Microscopic cross-sectional views of Hematoxylin & Eosin (Left, scale bar, 1 mm) and Masson Trichrome (Right, Scale bar: 50 μm) stained hearts isolated from Flnc icKO or Ctrl mice, 2 weeks or 6 weeks post TAM. n=3. (E) HW to BW ratio for icKO (n = 14) and Ctrl (n = 12) mice 2 weeks post TAM. (F) qRT-PCR analysis in Ctrl and icKO mouse hearts at 2 weeks post TAM. (G) Representative echocardiographic images of icKO and Ctrl mice at 2 and 4 weeks post TAM. (H-J) Echocardiographic measurements for Flncf/f and Flncf/f;Cre+ mice at baseline or 2, 4, 8 weeks post TAM or vehicle control. n = 8, 4, 15, and 5 respectively. (K) Diagram of intercalated disc (ICD), Z-disc, and costamere components. Proteins noted in red were upregulated, while those in black were unchanged, in Flnc icKO hearts. (L-P) Representative western blots and quantification of Integrin complex, DGC proteins, Z-disk protein and Desmin and ICD components in icKO and Ctrl hearts at 2 weeks post TAM (n=3). Other than FAK, δ-SAG, β-DSG, and Dystrophin which were not detected in our mass spectrometry studies, all other proteins tested in the western blot analysis have similar changes in mass spectrometry studies. (Q) qRT-PCR analysis for mRNAs of proteins examined in Ctrl and icKO mouse hearts at 2 weeks post TAM injection. (R) Functional enrichment analysis of proteins upregulated in icKO. For western blot analysis, GAPDH served as a loading control. The pixel density of each protein was normalized to the level of GAPDH. In qRT-PCR assays, mRNA levels of each gene were normalized to corresponding 18S levels, with icKO values being expressed as fold-change versus control. Data are represented as the mean ± SEM. *P < 0.05, by 2-tailed Student’s t test.