Abstract
A multiplex PCR system has been developed and optimized for rapid detection of the five main pathogens of bacterial pneumonia. The system can be expanded to analyze viral pathogens of pneumonia (DNA- and RNA-containing viruses), as well as those of a fungal nature.
Keywords: infectious pneumonia, multiplex PCR, diagnostics
INTRODUCTION
Infectious pneumonia is an acute inflammatory process of the lung tissue caused by a wide range of pathogens of bacterial, viral and fungal nature. Outbreaks of infectious pneumonia caused by SARS-CoV coronaviruses (2002–2003 yr), MERS-CoV (2012–2013 yr), SARS-CoV-2 (2019–? yr), Influenza A and B viruses, as well as bacterial pneumonia, can become a real social threat. Bacterial pneumonia is characterized by a mortality rate of about 15%, while mortality in cases of the severe course of community-acquired pneumonia (SAP) can reach 21–58% [1], which is comparable to those of SARS (so-called atypical pneumonia), MERS [2] and exceeds this indicator for CoViD-19 and influenza.
Specialized medical institutions that receive patients with a clinical diagnosis of pneumonia are faced with the problem of establishing the etiology of the disease and rapid identification of the pathogen, since often viral and bacterial pneumonia are characterized by a similar clinical picture. The situation sharply worsened with the onset of the CoViD-19 pandemic, which requires timely differential diagnosis of a secondary bacterial infection, which often arises as one of the complications of the course of the disease.
The correct selection of drugs and the patient’s treatment strategy depend on the timely diagnosis [3]. The current situation signals an urgent need to create express methods of differential diagnostics.
Multiplex PCR was developed for the species determination of the five main pathogens of bacterial pneumonia in humans. The system is focused on the use in clinical diagnostics to determine the etiology of the disease due to the ability to differentiate bacterial from viral and fungal pneumonia.
RESULTS AND DISCUSSION
One of the most common pathogens of bacterial pneumonia are representatives of several genera: Staphylococcus aureus, Pseudomonas aeruginosa, Haemophilus influenza, Legionella pneumophila, Klebsiella pneumonia and some others [4]. For these five bacterial species, genetic targets were selected and primers were designed to perform multiplex PCR. When designing the primers, they were guided by the requirement of species specificity and intraspecific conservatism of selected regions of genetic targets, which is necessary for reliable differential diagnostics, for example, the ebpS gene for identification S. aureus [5] and the gene sidA for L. pneumophila [6]. We took into account the need to obtain different lengths of PCR products for the convenience of identifying the pathogen during electrophoretic separation.
For each primer, a BLAST analysis procedure was performed, its physicochemical characteristics were determined, including testing for the presence of both intramolecular and intermolecular secondary structures.
The temperature-time profile of PCR was optimized using gradient PCR, as well as the composition of the buffer components and the concentration of each of the primers in the mixture. The specificity of primers to target and non-specific templates was determined experimentally (using the total genomic DNA of each of the tested bacterial strains), both in the mode of application of individual DNA templates and in the mode of DNA mixtures of several pathogens in a single tube. It was found that primers have high specificity in a mixture containing the DNA of various microorganisms and are able to identify solely their specific targets, without giving a false positive result in the presence of nonspecific DNA in the mixture.
The sensitivity of the test system was determined by DNA dilutions of each of the analyzed pathogens, which amounted to 102 to 103 copies of genomic DNA to the reaction tube, including the simultaneous introduction of several templates into the mixture.
The result of differential detection of the pathogen DNA in the sample by multiplex PCR is shown in Fig. 1.
Fig. 1.
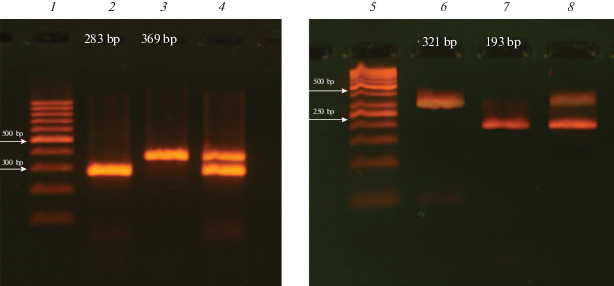
Determination of pathogen DNA by electrophoretic separation of PCR products. (1) GeneRuler 100 bp DNA length marker (Thermo Scientific, United States), (2) S. aureus, (3) L. pneumophila, (4) S. aureus + L. pneumophila, (5) GeneRuler 50 bp DNA length marker (Thermo Scientific, United States), (6) P. aeruginosa, (7) H. influenza, (8) P. aeruginosa + H. influenza. 4% agarose gel, ethidium bromide staining.
The developed system can be expanded to identify pathogens of pneumonia of a viral and fungal nature. Currently, it is planned to conduct tests on clinical samples, use labeled dNTP derivatives for subsequent analysis on biochips, and also develop a mathematical algorithm for calculating amplification signals. The system is suitable for diagnostic laboratories specializing in clinical analysis using PCR.
EXPERIMENTAL
Strains
In the work, decontaminated full-genome DNA of bacterial strains from the SSC Collection of Applied Microbiology and Biotechnology (Obolensk) was used. DNA was isolated from cell cultures using the CTAB method [7].
Primers
Multiple alignment of sequences of genomic targets was performed using ClustalW algorithm (www.clustal.org). The design of the primers was carried out using a network resource www.idtdna.com, analysis of specificity was performed using the BLAST algorithm (NIH, United States). The sequences, species specificity, genetic targets and length of PCR products for all primer pairs used are given below: S. aureus, ebpS gene, forward ebpS-f (5'-ACTCGACTGAGGATAAAGCGTCT-3'), reverse ebpS-r (5'-CCTCCAAATATCGCTAATGCACC-3'), product length 283 bp; L. pneumophila, sidA gene, forward sidA-f (5'-TTCCACTGGTGGGTGGGGTTTTG-3'), reverse sidA-r (5'-TCATGTTGGAGTTCTATGGCACG-3'), the length of the PCR product is 369 bp; H. influenza, gene fucK, forward fucK-f (5'-TGCTCACTCAACGCTTAACTGGT-3'), reverse fucK-r (5'-TTCTGGGCTAATGGTGTACGTAA-3'), the length of the PCR product is 193 bp; P. aeruginosa, gene oprL, forward oprL-f (5'-GCGTGCGATCACCACCTTCTACT-3'), reverse oprL-r (5'-TTCTTCAGCTCGACGCGACGGTT-3'), the length of the PCR product is 321 bp; K. pneumonia, gene rmpA, forward rmpA-f (5'-ATCAATAGCAATTAAGCACAAAAA-3'), reverse rmpA-r (5'-TCATAATCACACCCTTTAGGATA-3'), the length of the PCR product is 177 bp.
Multiplex PCR
The reaction mixture (30 μL) contained 1.5 units of Taq polymerase (Thermo Scientific, United States) in a buffer of the same company, dNTP mix at a concentration of 200 μM each, five pairs of specific primers and a full-genome bacterial template (or a mixture of bacterial DNAs). The reaction was carried out on a MiniCycler DNA Amplifier (MJResearch, United States) under the following conditions: 95°C for 5 min (initial denaturation); 36 cycles of 20 s at 95°C, 30 s at 66°C and 40 s at 72°C; final incubation for 5 min at 72°C. Gradient PCR and determining the sensitivity of the system using real-time PCR were performed on an IQ5 amplifier (Bio-Rad, United States). PCR products were separated on a 4% agarose gel, stained with ethidium bromide. The type of DNA analyzed was determined by the lengths of amplification products (Fig. 1).
FUNDING
The study was supported by the Russian Science Foundation (grant no. 20-14-00287).
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any research involving humans and animals as research objects.
Conflict of Interests
The authors declare no conflict of interest.
Footnotes
Abbreviations: SARS, severe acute respiratory syndrome; SAP, severe community-acquired pneumonia; CTAB, cetyl-trimethyl ammonium bromide.
Corresponding author: phone: +7 (495) 135-98-00; fax: +7 (495) 135-14-05; e-mail: lapa@biochip.ru.
REFERENCES
- 1.Sligl W.I., Marrie T.J. Crit. Care. Clin. 2013;29:563–601. doi: 10.1016/j.ccc.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. Viruses. 2019;11:E59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris, M., Clark, J., Coote, N., Fletcher, P., Harnden, A., McKean, M., and Thomson, A., Thorax, 2011, vol. 66, ii1eii23. 10.1136/thoraxjnl-2011-200598 [DOI] [PubMed]
- 4.Han Y.C., Woo J.H. Respirology. 1996;1:115–122. doi: 10.1111/j.1440-1843.1996.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Cao Y., Wang T., Dong Q., Li J., Niu C. Front. Microbiol. 2019;10:A222. doi: 10.3389/fmicb.2019.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabatabaei M., Hemati Z., Moezzi M.O., Azimzadeh N. Mol. Biol. Res. Commun. 2016;5:215–223. [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle J.J., Doyle J.L. Phytochem. Bull. 1987;19:11–15. [Google Scholar]


