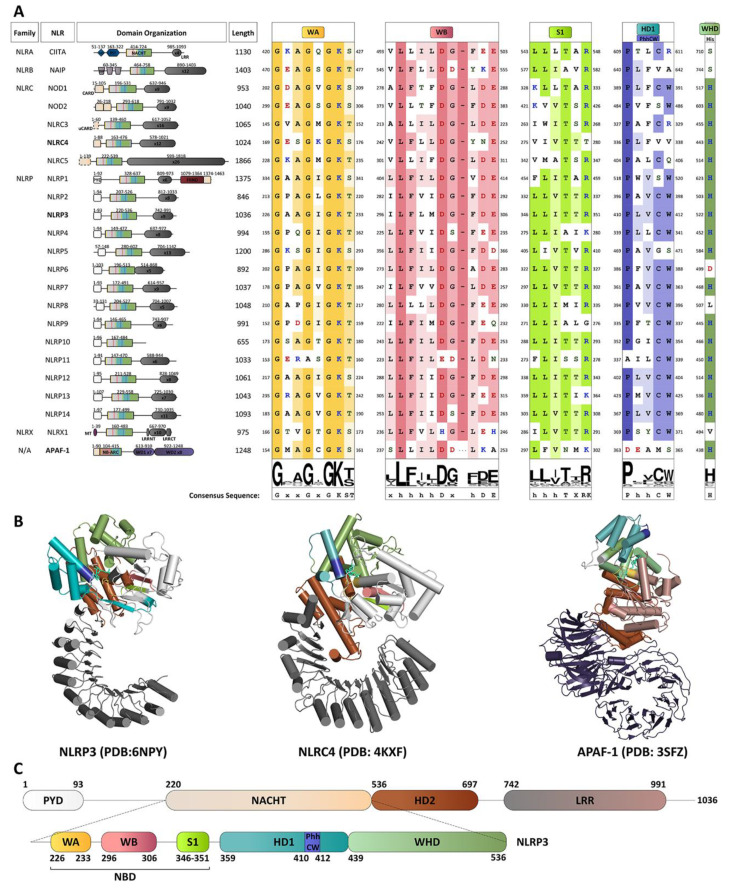
Figure 1.
Characterization of NLRP3 and the NLR family. (A) Multiple sequence alignment of conserved features critical for catalytic activity in the 22 NLR proteins and APAF-1 NBD motifs. The NLR family members are subdivided based on the presence of one of four N-terminal effector domains: NLRAs with an acidic domain (AD), NLRBs with a Baculovirus IAP repeat (BIR), NLRCs with a caspase recruitment domain (CARD), or NLRPs with a pyrin domain (PYD). The naming system approved by the HUGO Gene Nomenclature Committee (HGNC) is used [10]. Domain identities and configuration for NLR proteins are displayed to scale with boundaries numbered as per UniProtKB accession numbers: CIITA, P33076; NAIP, Q13075; NOD1, Q9Y239; NOD2, Q9HC29; NLRC3, Q7RTR2; NLRC4, Q9NPP4; NLRC5, Q86WI3; NLRX1, Q86UT6; NLRP1, Q9C000; NLRP2, Q9NX02; NLRP3, Q96P20; NLRP4, Q96MN2; NLRP5, P59047; NLRP6, P59044; NLRP7, Q8WX94; NLRP8, Q86W28; NLRP9, Q7RTR0; NLRP10, Q86W26; NLRP11, P59045; NLRP12, P59046; NLRP13, Q86W25; and NLRP14, Q86W24. Multiple sequence alignments were created with Jalview [16] and MUSCLE [17] with default options and used to generate sequence conservation logos shown below each motif [18]. These indicate sequence conservation amongst NLRs and APAF-1, with the height of the stack indicating overall sequence conservation at that position in the alignment, while the height of letters within the stack indicates the relative frequency of each amino acid at that position. Amino acid class in the alignment is denoted by coloured font; where hydrophobic residues (A,I,L,M,F,W,V,C,G,P) are coloured in yellow; polar residues in green; polar (S,T,Y,N,Q) in green; acidic (D,E) in red; and basic (K,R,H) in blue. Conservation within each motif is denoted by background shading, where no color indicates <30% conservation, and increasing colour intensity indicates conservation of residue type from 30–60%, 60–80% or >80%. (B) Cartoon representation of NLR proteins NLRP3 and NLRC4 as well as APAF1, a close structural homolog. Domains and motifs are coloured as in (A). (C) Schematic representation of human NLRP3 showing important domains and motifs. The labelled domain boundaries were defined as per UniProtKB accession Q96P20-1. Important motifs within the NACHT domain were identified based on previous definitions. PYD, pyrin domain; NBD, nucleotide-binding domain; HD1, Helical domain 1; NACHT, ATPase domain named after its discovery in NAIP, CIITA, HET-E and TP1 apoptosis regulator proteins; WHD, Winged helix domain; HD2, Helical domain 2; WA, Walker A motif; WB, Walker B motif; S1, Sensor 1 motif; HD1: Helical domain 1; xVP, NLR xVP motif; LRR, Leucine rich repeat; AD, Acidic domain; PST, Proline-serine-threonine-rich domain; CARD, CAspase recruitment domain; uCARD, Untypical CARD domain; FIIND, domain with function to find; MT, Mitochondrial-targeting sequence; LRRNT, LRR N-terminal domain; LRRCT, LRR C-terminal domain; NB-ARC, Nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4; and WD, WD40 repeats. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.).