Table 1.
Data types used and/or produced in the context of taxonomy, currently or potentially in the future, their predicted storage requirements and main issues to be solved to allow their efficient storage and reuse
| Current use in alpha-taxonomy | Potential and prospective use in taxonomy | Storage requirements (per specimen) | Established specialized repositories | Issues and gaps | |
|---|---|---|---|---|---|
| Regular images (e.g., .jpeg, .pdf, .tiff) | Regularly used | Images of different kinds will continue to be a main workhorse of taxonomic description and identification; new perspectives by machine-learning character extraction | Moderate to very high, depending on image quality and quantity | Yes (many specialized and generalist repositories will accept images) | Images produced in taxonomic revisions are rarely submitted to repositories; images are often not linked to specimen identifiers |
| High-resolution images (stacks etc.) (e.g., .tiff) | Increasingly used, e.g. in insects | As with regular images | High to very high | Yes (many specialized and generalist repositories will accept images) | As with regular images |
| Annotated images | Very rarely used | Documentation of morphology of small-sized organisms (e.g., on a microscopic slide) | High to very high | Only few specialized repositories | Requires development of standards for repositories and submitters |
| 3D microCT, photogrammetry, and laser scanners (e.g., stack of .tiff, polygon mesh such as .ply, .bend, .obj) | Used rarely but regularly, especially in vertebrates. Increasing use in invertebrates. | High importance to visualize internal features of an organism or 3D morphometrics, key method in cyberspecimen approaches | High to very high, depending on storage modality (e.g., polygon mesh vs. raw data) and level of resolution | Yes, several | Requires development of standards for repositories and submitters.See commentary by Hipsley and Sherratt (2019). |
| DNA sequences (Sanger) (e.g., .fasta, .fastq, .gb) | Regularly used for most organism groups, almost omnipresent in mycological taxonomy | DNA barcodes will continue to drive species identification and discovery, multigene phylogenies important for inferring relationships | Very low to low depending on the number of loci sequenced | Yes, several very well established ones | Sequences deposited in databases are not always curated postsubmission, leading to mismatches after taxonomic changes. |
| RNAseq (raw) (e.g., .fastq) | Not used | Potentially useful after read mapping and variant calling, but currently rarely used. | High | Yes (e.g., Sequence Read Archive) | No issues |
| RNAseq (assembly) (e.g., .fasta) | Very rarely used | Valuable source of sequences for phylogenomics and species delimitation | Low | Yes (e.g., Transcriptome Shotgun Assembly Sequence Database) | Assemblies are often not submitted to repositories, although they could be a valuable source of sequences for machine-learning species discovery pipelines |
| Amplicon (raw) (e.g., .fastq) | Not used | Not straightforward; requires filtering and preprocessing | High | Yes (e.g., Sequence Read Archive) | No issues |
| Amplicon (consensus OTUs) (e.g., .fasta) | Not used | Metabarcoding data helps ascertaining distribution and ecology of taxa | Low | Not really. Sequences 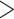 200 bp could be submitted to GenBank. 200 bp could be submitted to GenBank. |
OTU consensus sequences from metabarcoding studies are in most cases not submitted to a repository, but could be important for DNA-based assessments of distribution of taxa; targeted and searchable repositories do not exist (GenBank does not accept sequences <200 bp) |
| Bait capture—raw (e.g., .fastq) | Not used | Only usable after assembly | High | Yes (e.g., Sequence Read Archive) | No issues |
| Bait capture—assembled (e.g., .fasta) | Rarely used (e.g., sequencing of historical types) | Very valuable source of sequences for phylogenomics and species delimitation—next-generation DNA barcoding | Low to moderate | Yes, well established, same ones as for Sanger sequences | Similar as for Sanger sequences |
| Genomes—raw (e.g., .fastq) | Not used | Only usable after assembly | Very high | Yes (e.g., Sequence Read Archive) | Similar as for Sanger sequences |
| Genomes—assembled (e.g., .fasta) | Very rarely used | Valuable source of sequences for phylogenomics and species delimitation | High | Yes | Similar as for Sanger sequences |
| Maldi-TOF (e.g., .raw, .mzXML, .mzML) | Sometimes used in mycology; commonly in prokaryotes. | Useful for chemotaxonomic approaches | Moderate to very high, depending of storage of spectra vs. raw data. | No | Requires development of standards for repositories and submitters |
| Near-infrared spectroscopy (e.g., .snirf, .csv, .spc, and many others) | Not used | Possibly useful for “metabolomic barcoding” | Moderate | No | Requires development of standards for repositories and submitters |
| GC-MS/ (e.g., .raw, .cdf, .D, .mzxml) | Sometimes used in mycology; commonly in prokaryotes. | Useful for chemotaxonomic approaches, e.g., fatty acid profiling in yeasts, and in bacterial taxonomy | Moderate to very high, depending of storage of spectra vs. raw data. | No | Requires development of standards for repositories and submitters; reference databases do exist. |
| NMR/TLC/HPLC (e.g., .raw , .data, ..cdf) | Rarely used (e.g., TLC and HPLC in lichenology) | Possibly useful for chemotaxonomic approaches | Moderate to very high, depending of storage of spectra vs. raw data. | No | Requires development of standards for repositories and submitters |
| Sounds (e.g., .wav, .mp3) | Regularly used in sound-producing animals | Very useful for species delimitation of sound-producing animals | Moderate to high, depending on file format and sound duration | Yes | Most repositories do not feature user-friendly submission procedures and often data are not open access |
| Videos (.avi, .mov, .mp4) | Very rarely used (e.g., to document specific behavior) | Limited value | Moderate to very high, depending on definition and duration | Yes | Extend image databases to accept videos if linked to specimens |
| Measurements (e.g., .csv, .xls. .txt) | Regularly used | Very useful basic data for diagnosis and identification of species | Very low | No | Requires development of standards for repositories and submitters |
| 2D geometric morphometric data sets (e.g., .csv) | Very rarely used | Increasingly used for resolving species complexes | Very low to low | Yes | No issues |
| 3D geometric morphometric data sets (e.g., .csv) | Very rarely used | Increasingly used for resolving species complexes, especially in combination with microCT scans | Very low to low | Yes | No issues |
Note: Note that the second column specifically focuses on the current use of data types in alpha-taxonomic studies (mostly based on our survey reported below), not other taxonomy-related activities such as species identification or phylogenetics. Storage capacity required per specimen: very low (<0.1 MB), low (0.1–1 MB), moderate (1–10 MB), high (10–100 MB), and very high (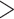 100 MB).
100 MB).
