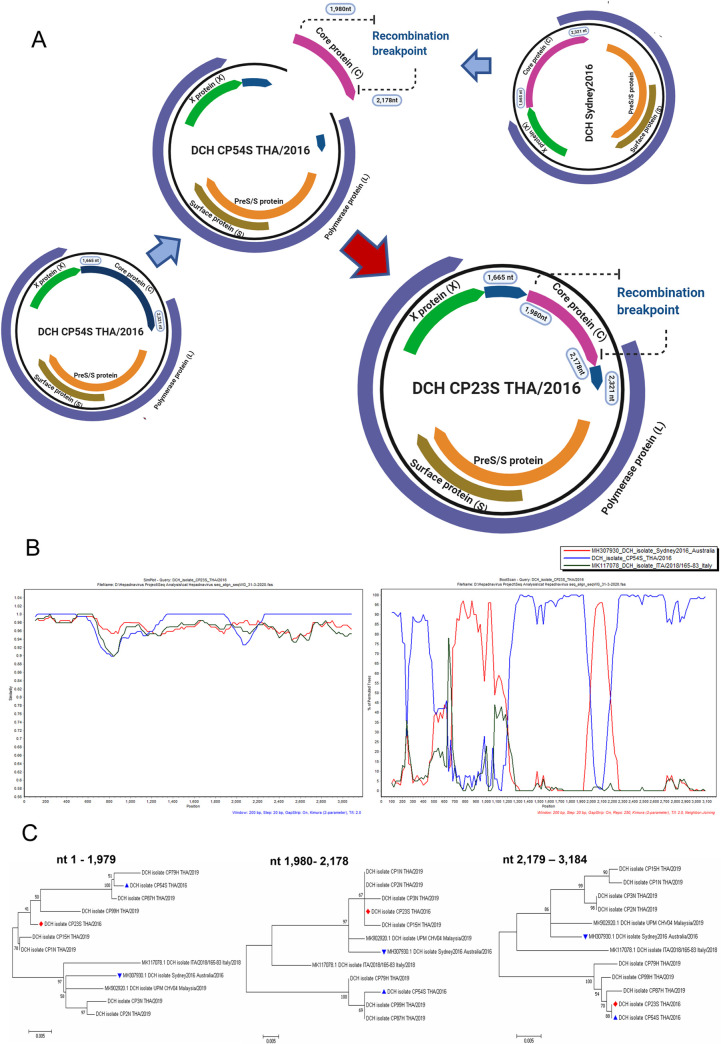
Fig 2. Schematic diagram of the recombinant DCH CP23S THA/2016 strain.
(A) The DCH CP54S THA/2016 isolated in this study and DCH Sydney2016 (MH307903) from Australia served as the putative major and minor parents. (B) The potential recombination event was detected in the core protein gene and was supported by similarity and bootscan analysis, which indicated that DCH CP54S THA/2016 (blue line) served as the main template of the whole DCH genome, and then the genome sequence was replaced by the sequence from the DCH Sydney2016 at most of the C gene. The DCH CP23S/2016 isolate served as the query. The y-axis indicated the percentage of nucleotide identity and permutated trees for the similarity plot and boot scanning, respectively, within a 200 bp-wide window with a 20-bp step size between plots. (C) The ML phylogenetic trees of the recombinant DCH23S THA/2016 strains (●) and its major (▲) and minor (▼) putative parent strains over three different segments. Bootstrap (1000 replications) values over 50% are shown for each node.