Figure 1. Overview of bioinformatic workflow.
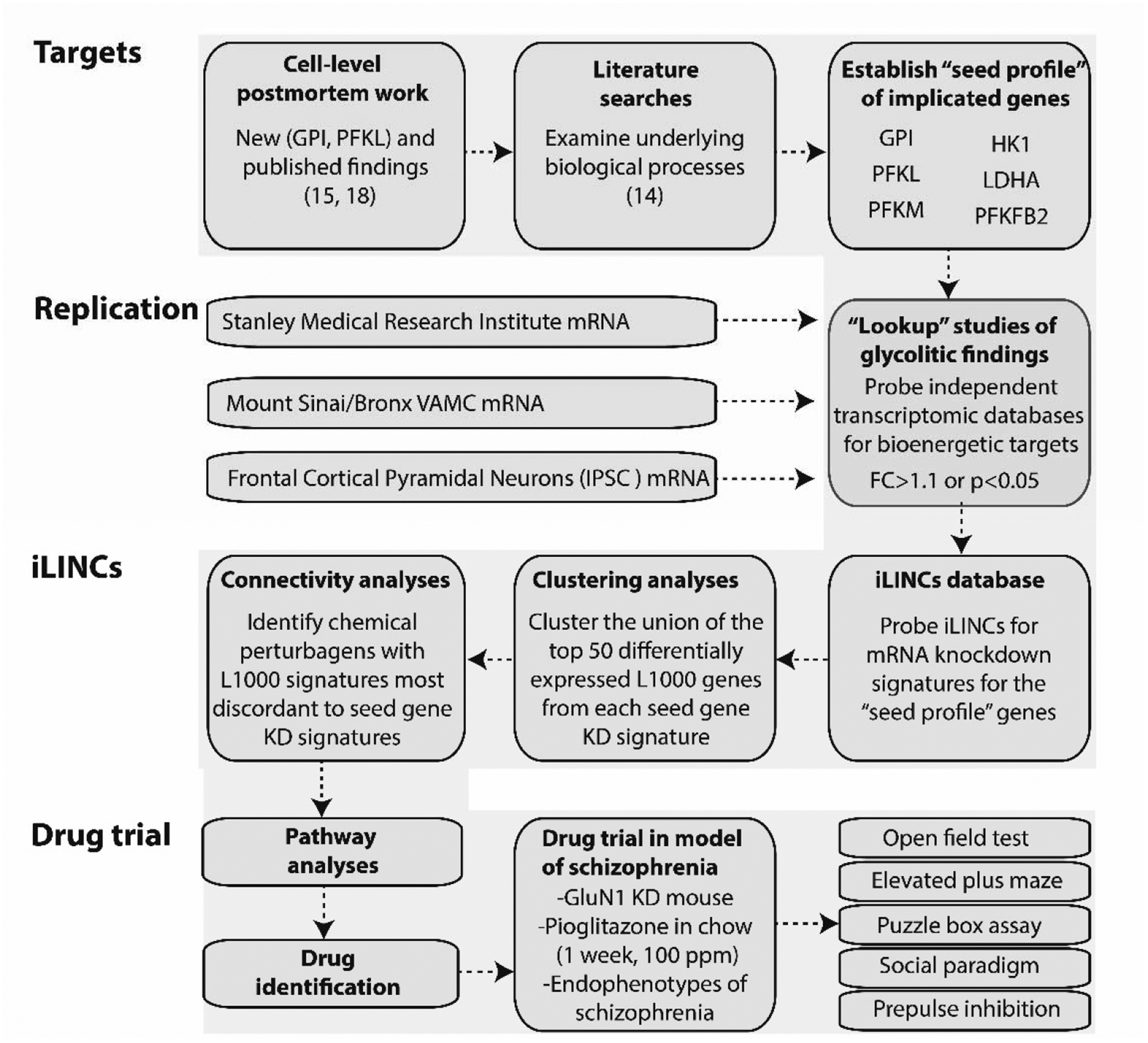
We began by performing literature searches and further characterizing cell-level postmortem bioenergetic abnormalities in schizophrenia. Based on these findings, we established a bioenergetic seed profile containing 6 glycolytic genes implicated in schizophrenia (“seed genes”). To replicate our bioenergetic findings in schizophrenia, we probed a publicly available microarray database (Stanley Medical Research Institute Online Genomics Database), microarray data from the DLPFC of control and schizophrenia subjects (Mount Sinai School of Medicine, Bronx VAMC mRNA dataset), and RNAseq data from frontal cortical neurons of a patient with schizophrenia and DISC1 mutation (75). Next, we used iLINCS (http://ilincs.org) to retrieve knockdown signatures for each seed gene individually. With the goal of identifying panels of correlated transcriptomic changes in our seed gene KD signatures, we used iLINCS to perform unsupervised clustering of the top 50 differentially expressed L1000 genes from our 6 seed gene KD signatures. The panels of clustered differentially expressed L1000 genes were carried forward to Enrichr analyses. We also performed iLINCS connectivity analyses and identified peroxisome proliferator-activated receptor (PPAR) agonists as promising therapeutic targets with the potential to “reverse” glycolytic deficits as well as the associated changes in the L1000 genes. Finally, we administered the PPAR agonist pioglitazone to the GluN1 knockdown model of schizophrenia an examined endophenotypes of schizophrenia. Phosphofructokinase liver type (PFKL); PFK muscle type (PFKM); Glucose-6-phosphate isomerase (GPI); 6-phosphofructo-2-kinase (PFKFB2); lactate dehydrogenase A (LDHA); hexokinase 1 (HK1); fold change (FC); inducible pluripotent stem cells (IPSC); knockdown (KD).
