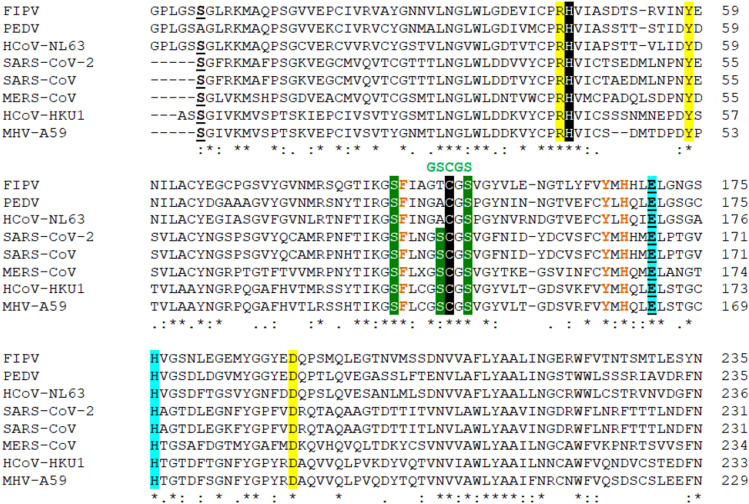
Fig. 2.
The multiple sequence alignment of 3CLpro of CoVs of different origin (using the Clustal Omega program). The Cys145 of SARS-CoV-2 3CLpro that interact with the compounds 11a and 11b is shown in black background along with the Cys145-His41 catalytic dyad highly conserved in 3CLpro from CoVs [42, 44, 59, 65, 66]; in green are marked the cluster of Ser with high affinity for small molecule inhibitors [44, 55]; in green it is shown the GSCGS motif essential for starting the catalysis; in light blue are marked the Glu-His residues critical for substrate binding by means of steric effect; in yellow background is marked the triad Arg-Tyr-Asp that forms a partial negative charge cluster that, by a conserved water molecule, mediates the interaction with Cys-His catalytic dyad; in brown are shown the residues involved in the glutamine substrate recognition—the conserved His and the conserved Tyr and Phe that interacts with by the phenolic hydroxyl group with His and employs a steric effect to restrain the rotation of His, respectively; in bold-underline are marked the Glu and Ser residues that are demonstrated to be essential in the dimer interactions in SARS-CoV-2 [60]; with dot “.” are marked the semi-conservative replacements; with colon “:” are marked the conservative replacements; with “*” are marked the identities of the residues