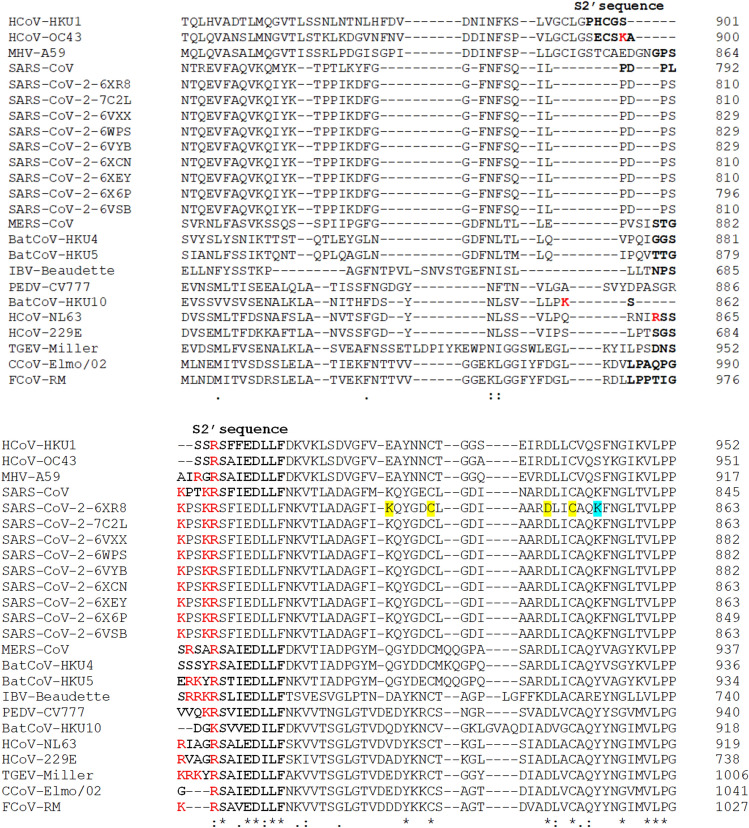
Fig. 7.
The multiple sequence alignment of spike (S) S2′ cleavage site sequences using the Clustal Omega program. The S1/S2 sequences are bolded and the basic arginine and lysine residues are marked in red [93]; in yellow background are marked the cysteine residues that form an internal disulfide bond (C840 and C851) and the residues that form a salt bridge that reinforces the previous disulfide bond (K835-D848); in light blue is shown the lysine (K854) that form a salt bridge with the aspartic acid (D614) (not shown) [40, 107–109]