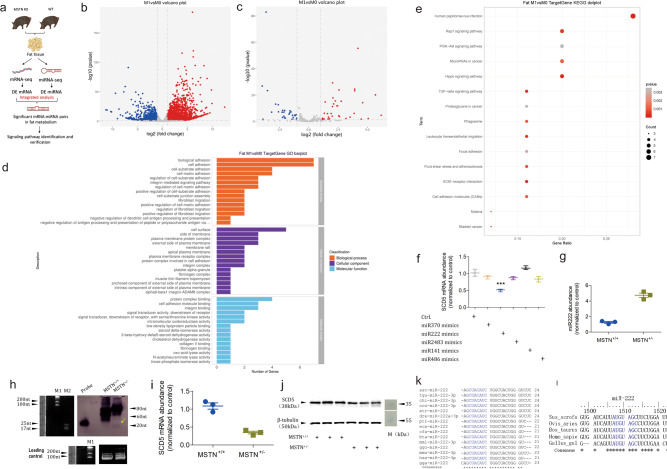
Fig. 2. Identification of miRNA-mRNA regulatory pairs potentially involved in fat deposition.
a The schematic of mRNA and miRNA deep sequencing and bioinformatic analysis aimed at identification of potential signaling pathways. b Volcano plot of the differentially expressed mRNAs between MSTN-KO and WT pigs. c Volcano plot of the differentially expressed miRNAs between MSTN-KO and WT pigs. M0 and M1 represent WT and MSTN-KO pigs, respectively. d GO barplot of differentially expressed mRNAs involved in lipid metabolism. e KEGG dotplot of differentially expressed mRNAs involved in lipid metabolism. f qPCR analysis of SCD5 mRNA abundance in response to treatments with five miRNAs. g qPCR analysis of miR222 abundance in the subcutaneous fat tissue of WT and MSTN-KO pigs (n = 3 biologically independent samples, respectively). h Northern blotting of miR222 in WT and MSTN-KO pigs. The yellow arrowhead denotes the position of miR222. Small RNAs were loaded in equal amounts. Probe against miR222 was used as positive control for DIG colorization. M1 and M2 are RiboRuler Low Range RNA Ladder (SM1831, ThermoFisher Scientific) and microRNA Marker (N2102, NEB). i qPCR analysis of endogenous SCD5 abundance in the subcutaneous fat tissue of WT and MSTN-KO pigs (n = 3 biologically independent samples, respectively). j Western blotting of the endogenous SCD5 in the subcutaneous fat tissue of WT and MSTN-KO pigs (n = 3 biologically independent samples, respectively). M is the Page Ruler Prestained Protein Ladder 26616 (ThermoFisher Scientific). k Evolutionary conservation of miR222 in multiple species. Red letters represent the seed sequence of miR222. “*” Represents the evolutionary consensus sequence of miR222. l Binding sites of miR222 on the 3′UTR of SCD5 in five animal species. Red letters represent the annealing region of seed sequence of miR222. “*” Shows the consensus sequence of 3′UTR of SCD5 in the five animal species.