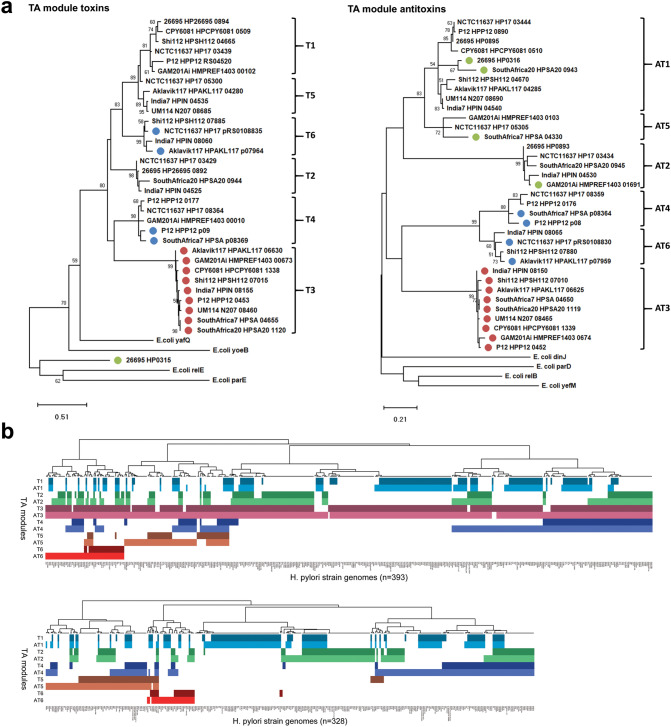
Figure 1.
Phylogeny, prevalence and sequence conservation of H. pylori YafQ-family Type II toxin–antitoxin modules. (a) Phylogenetic analysis of H. pylori YafQ-family Type II TA module toxin and antitoxin nucleotide sequence variation using the Maximum Likelihood method. Toxin and antitoxin clades comprising representative TA subset sequences are labelled T1–T6 and AT1–AT6 respectively and include Type II antitoxin sequences associated with the non-Type II vapD toxin locus. TAs clearly associated with mobile elements or the vapD loci are indicated by coloured circles (blue; plasmid, red; tfs4 ICE, green; vapD loci). The percentage (> 50%) of replicate trees in which the associated sequences clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. (b) Prevalence of Type II TA modules in a global population of H. pylori strains. Hierarchical clustering indicates the distribution and co-occurrence of TA toxin subsets in both the presence (top, 393 strains) and absence (bottom, 328 strains) of the tfs4 ICE encoded Clade T3/AT3 TA module.