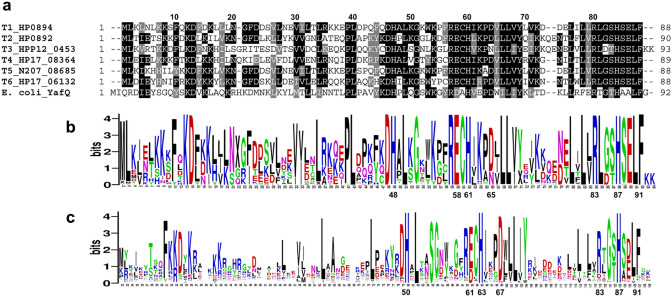
Figure 4.
Sequence conservation of YafQ-family toxins. (a) Multiple sequence alignment of representative H. pylori Clade T1–T6 YafQ-family toxin sequences with E. coli YafQ. Numbering above the alignment refers to HP0894 sequence. WebLogos generated from multiple alignment of (b) Clade T1–T6 consensus sequences (n = 701) from a geographically diverse collection of H. pylori strains and (c) related Type II toxins from 318 other genera of bacteria. Sequences from multiple species from the majority of genera were included in the alignment to capture the full extent of toxin sequence diversity (n = 934 consensus sequences, Supplementary File S1). Numbering below each WebLogo refers to the position of conserved active-site residues His50, His63, Asp67, Arg83, His87 and Phe91 of E. coli YafQ (c) and the corresponding position of equivalent TfiT residues (b). In each logo, the height of individual amino acid letter codes corresponds to the prevalence of particular residues at each position in the protein sequence. A height of 4.2 bits is indicative of 100% conservation.