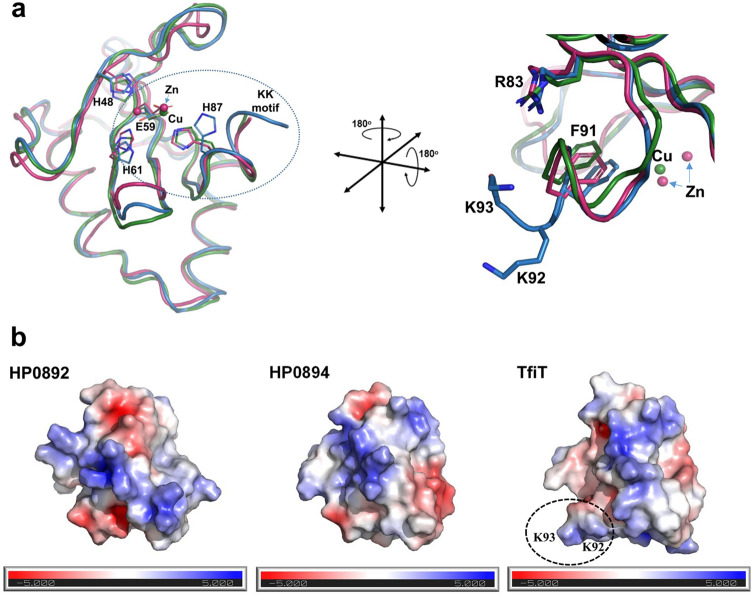
Figure 6.
Structural homology of TfiT with genomic toxins HP0892 and HP0894. (a) Superposition of the crystal structures of Cu-bound HP0894 (PDB: 4LSY, green), Zn-bound-HP0892 (PDB: 4NRN, pink) and modelled TfiT (blue) highlighting side-chains of conserved active site residues important for acid–base catalysis during mRNA hydrolysis, shown in stick form, relative to the TfiT C-terminal lysine motif. (b) The C-terminal lysines of TfiT extend the electrostatic interface for substrate attraction and interaction. Surface representations of HP0894 (PDB: 4LSY), HP0892 (PDB: 4NRN) and modelled TfiT showing the electrostatic potential surface in the same orientation plane as (a), right. The accessible surface area is coloured according to calculated electrostatic potential from − 5 kBT/e (red) to + BT/e (blue).