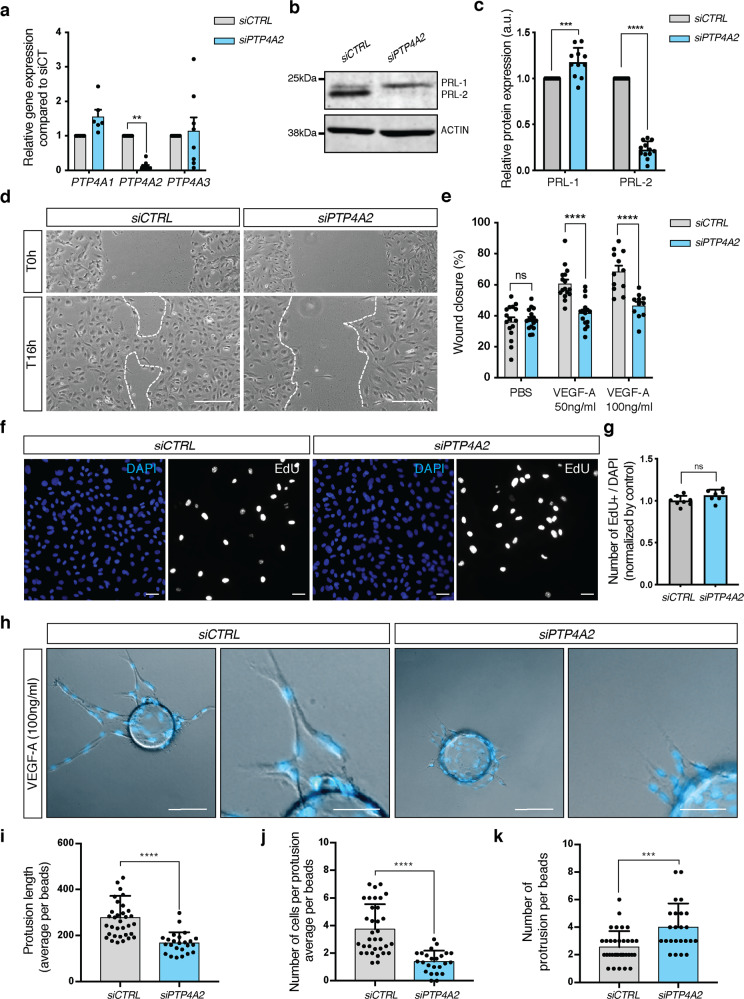
Fig. 5. Effect of PRL-2 silencing on endothelial cell migration and sprouting in vitro.
a qPCR analysis in HUVEC cells after CTRL or PTP4A2 siRNA treatment (at least n = 6 independent experiments; two-way ANOVA: *p value < 0.05). b Western blot in HUVEC cells, and c PRL-1 and PRL-2 protein quantification of western blot shown in b after CTRL or PTP4A2 siRNA treatment (n = 10 independent experiments; two-way ANOVA: ***p value < 0.001, ****p value < 0.0001). d Scratch wound assay performed on HUVEC monolayer in the presence of VEGF-A (50 ng/ml) after CTRL or PTP4A2 siRNA treatment. Images at 0 and 16 h after scratch. e Analysis and quantification of the wound closure in HUVEC cells after CTRL or PTP4A2 siRNA treatment in the presence (50 and 100 ng/ml), or absence of VEGF-A (PBS). (Each individual data point represents a biological replicat from n = 3 independent experiments, two-way ANOVA: ****p value < 0.0001, ns nonsignificant p value > 0.05). f, g EdU incorporation assay (8 h) on HUVEC cells after CTRL or PTP4A2 siRNA treatment (Mann–Whitney U test: ns nonsignificant p value > 0.05). Each individual data point represents a biological replicat from n = 3 independent experiments. h HUVEC sprouting assay embedded in 3D fibrinogen gel and stimulated with VEGF-A (100 ng/ml) after CTRL or PTP4A2 siRNA treatment. Quantification of i protrusion length, j number of cells per protrusion, k number of protrusions per beads. Results were expressed as average/beads. Each individual data point represent a beads; n = 3 (Mann–Whitney U test: ***p value < 0.001, ****p value < 0.0001). Scale bars: d 250 μm, f 50 μm, h 100 and 50 μm. Error bars represent mean ± s.e.m.