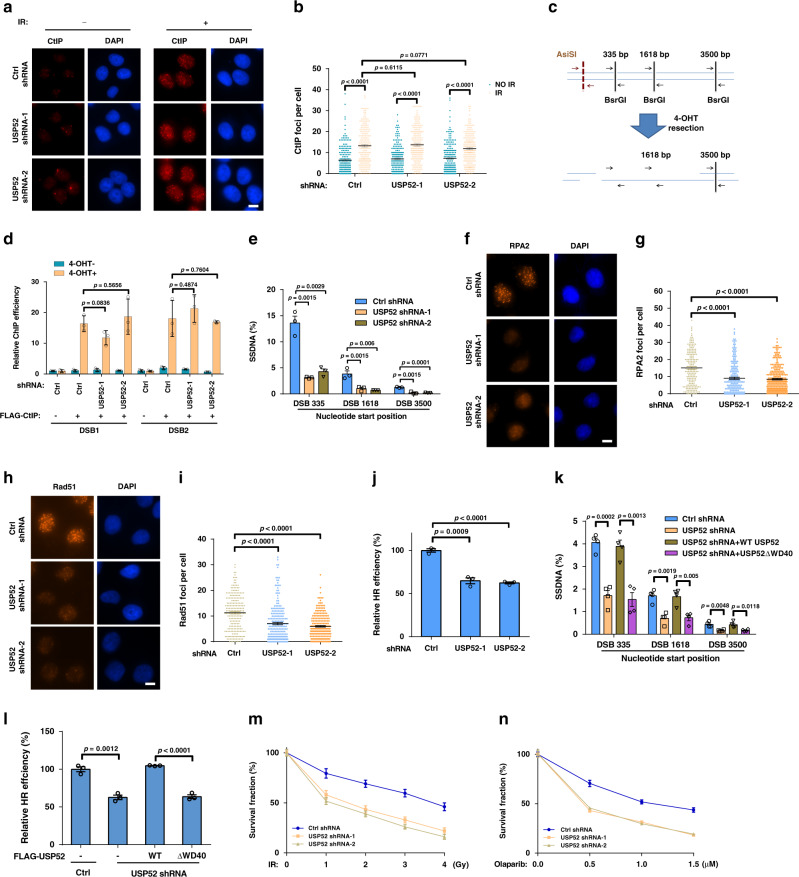
Fig. 2. USP52 promotes DNA end resection and HR repair.
a, b, f–i Representative pictures (a, f, h) and quantifications (b, g, i) of CtIP (a, b), RPA2 (f, g) and Rad51 (h, i) foci in control or USP52-depleted U2OS cells when treated with 5 Gy IR for 3 h, 2 Gy IR for 2 h or 5 Gy IR for 5 h, respectively. Data are representative of three independent experiments. Each dot represents a single cell, and more than 200 cells were counted in each group for this experiment. Error bars represent SEM from this experiment. Scale bar, 10 μm. c Illustration of the designation of Taqman qPCR primers and probes (black arrows) for measuring resection at sites adjacent to the AsiSI sites (red arrows). The primer pairs are across BsrGI restriction sites. All Taqman probes are designed at either side of the restriction site. d Control or USP52-depleted ER-AsiSI U2OS cells transfected with FLAG–CtIP were added with or without 4-OHT. ChIP assay was then performed using FLAG antibody. e The genomic DNA extracted from control or USP52-depleted ER-AsiSI U2OS cells were digested with BsrGI. DNA end resection adjacent to DNA double-strand break sites was measured by qPCR assay. j The HR-mediated DSB repair efficacy of control and USP52 depletion in HEK293T cells were analyzed using HR reporter. Data are presented as mean values ± SEM from three independent experiments. k Control or USP52-depleted ER-AsiSI U2OS cells transfected with indicated constructs were used for DNA end resection assay. Data are presented as mean values ± SEM from four independent experiments. l The efficacy of HR in control or USP52-depleted HEK293T cells transfected with indicated USP52 constructs were analyzed using GFP reporter assay. m, n Control or USP52-depleted U2OS cells were exposed to the indicated dose of IR (m) or PARPi (n) for 2 weeks, colony formation assay was then performed to detect the survival rate of cells. d, e, j, l–n Data are presented as mean values ± SEM from three independent experiments.