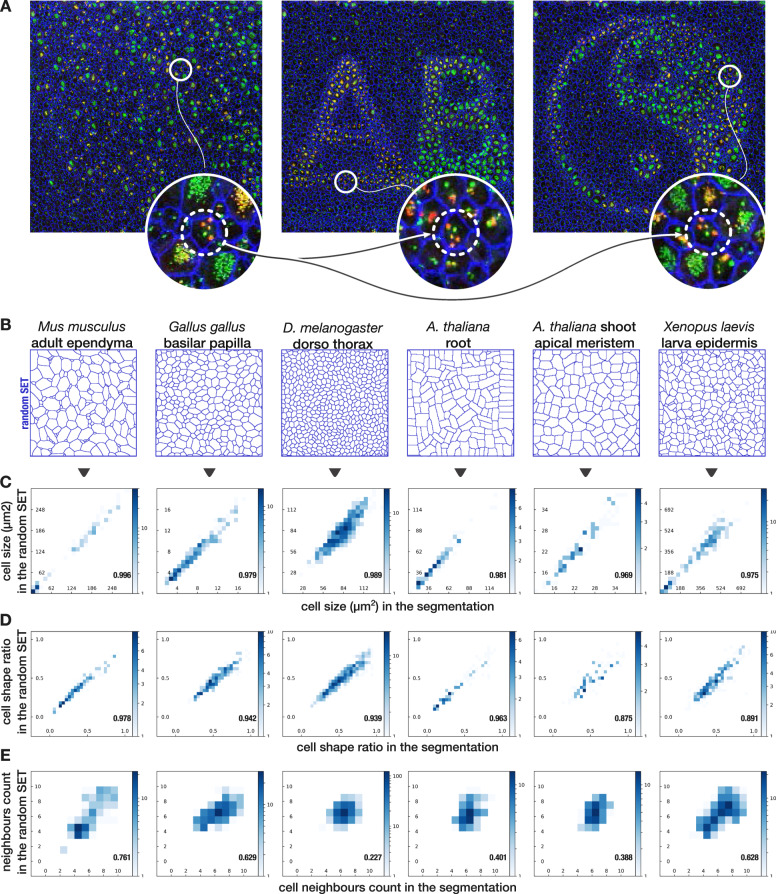
Fig. 2. Generation of alternative SET preserves single-cell size and shape ratio while expectedly breaks local cell organization.
a Left: an image of P3 mice lateral ventricular surface where borders (ZO1 antibody, blue), centrioles (Centrin2-GFP mice, green), and procentrioles (Sas6 antibody, red) are labeled. Middle and right: two alternative SET with the exact same 2822 cells taken from a and artificially moved to specified locations for illustrating the capability of SET. Only the positional parameters are modified so that cell shapes remain approximately the same. All cells were moved except those touching the borders of the image (as they are incomplete). Zoom on the same cell in the three images shows that intracellular content of each cell could also be transported to its new location using barycentric coordinates relative to the cell contour (see “Methods” for details). Interestingly, single-cell properties such as size and cell minor axis to major axis ratio is preserved while neighborhood is expectedly broken (Supplementary Fig. 3). b The same process is applied on all images presented in Fig. 1e but this time, location and orientation are randomly initialized. c–e 2D histograms showing the comparison of quantitative features computed for each cell both on the segmentation and on the random SET. Pearson correlation coefficient computed on all the cells except the border cells: c Cell size correlation. d Cell minor axis to the major axis ratio correlation. e Cell neighbors count correlation. The correlation of this last feature is expectedly broken as it depends on the surrounding of each cell which is randomly shuffled by the process. For all plots N = 1 image.