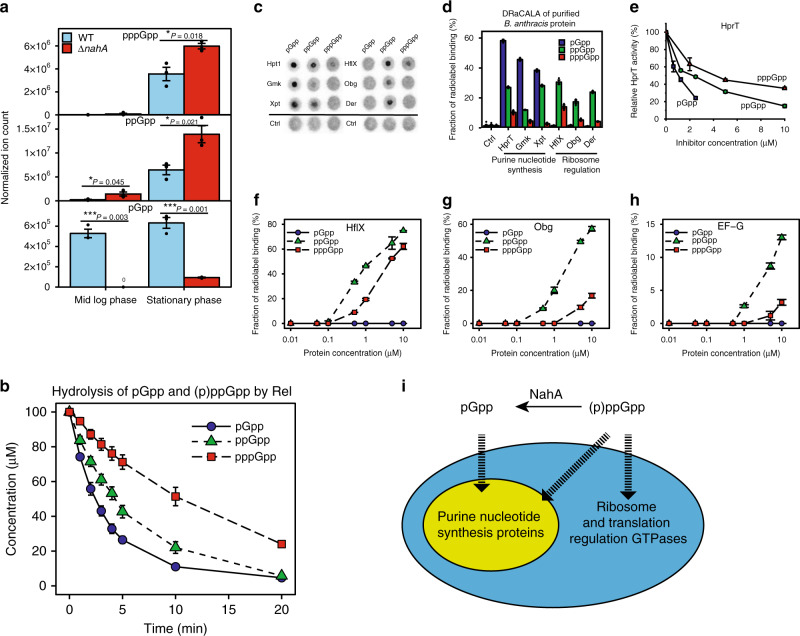
Fig. 3. pGpp is produced by NahA in vivo and has a distinct binding spectrum compared to (p)ppGpp.
a LC-MS analyses of pGpp, ppGpp, and pppGpp of wild type and ΔnahA in log phase and stationary phase. Normalized ion count is ion count per OD600nm per unit volume of the culture. Error bars represent standard errors of the mean of three biological replicates. A two-tailed two-sample equal-variance Student’s t test was performed between samples indicated by asterisks. Asterisks indicate statistical significance of differences (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). b In vitro hydrolysis of pGpp, ppGpp, and pppGpp by B. subtilis Rel. For each nucleotide, 200 nM Rel was incubated with a mix of 100 μM unlabeled alarmone and a small amount of 5′-α-32P-labeled version. The degradation of the alarmone was analyzed by TLC to measure the decreased radioactivity. Error bars represent standard errors of the mean of three replicates. c DRaCALA of purified His-MBP-tagged B. anthracis proteins (1 μM) with <0.1 nM of identical amount of 5′-α-32P-labeled pGpp, ppGpp, and pppGpp. d Quantification of DRaCALA in c. Error bars represent standard errors of the mean of three replicates except for control group (six replicates). e HprT enzymatic activities in the presence of indicated concentrations of pGpp, ppGpp, and pppGpp. The reaction was performed with 20 nM HPRT, 1 mM PRPP, and 50 μM guanine14. Error bars represent standard errors of the mean from three replicates. f–h Titrations of B. anthracis GTPases and quantification of their binding to pGpp, ppGpp and pppGpp using DRaCALA: GTPases HflX (f), Obg (g), and translation elongation factor G (EF-G) (h). Error bars represent standard errors of the mean of three replicates. i Schematic showing the relationship between pGpp-binding targets and (p)ppGpp-binding targets.