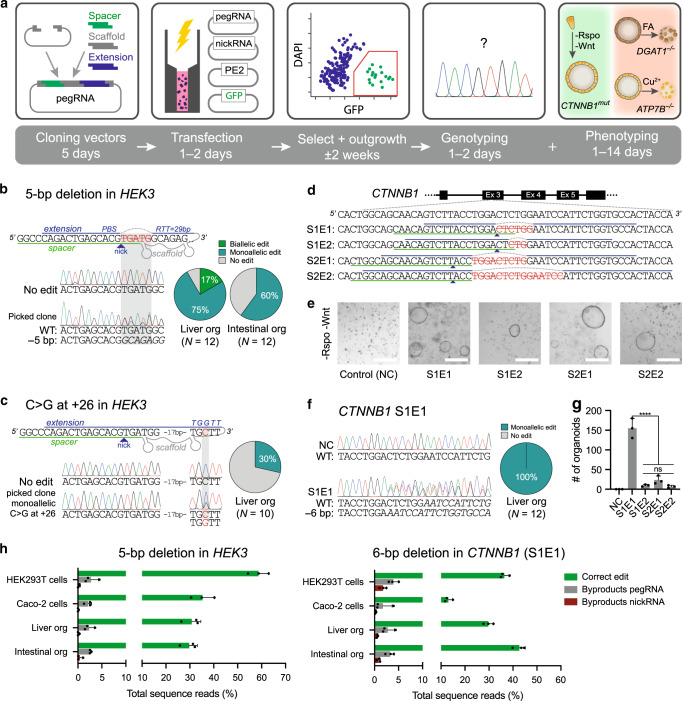
Fig. 1. Prime editing efficiently creates deletions and point mutations in organoids.
a Schematic overview of the workflow and timeline to generate precise mutations in organoid cells using prime editing (PE3). b pegRNA design, Sanger validation in a clonal organoid with monoallelic edit, and editing efficiency of a 5-bp deletion in HEK3 in liver and intestinal organoids. c pegRNA design, Sanger validation of monoallelic edit, and editing efficiency for a C → G substitution in HEK3 in liver organoids. Nicking sgRNA at +90 used in b and c not shown. d pegRNA designs for the generation of in-frame deletions in the β-TrCP region of CTNNB1. Nicking sgRNA at +86 (S1) or +93 (S2) not shown. e Brightfield images of liver organoid cells transfected with plasmids from d after Rspo1 withdrawal for 2 weeks. White scale bars are 500 µm. f Sanger validation of precise 6-bp deletions in all picked clones from CTNNB1 pegRNA S1E1 that continue growing in -Rspo1 conditions. g Quantification of organoid outgrowth in e. p < 0.0001 in a one-way ANOVA with Holm–Sidak correction. h Comparison of editing efficiencies and generation of unwanted byproducts in different cell types by high-throughput sequencing (HTS). Only transfected cells (GFP+ sorted) were used for HTS. Data are represented as mean values ±S.D. of three independent experiments g or biological replicates h. FA fatty acids, PBS primer binding site, RTT reverse transcriptase template, NC negative control. Source data are provided as a Source Data file.