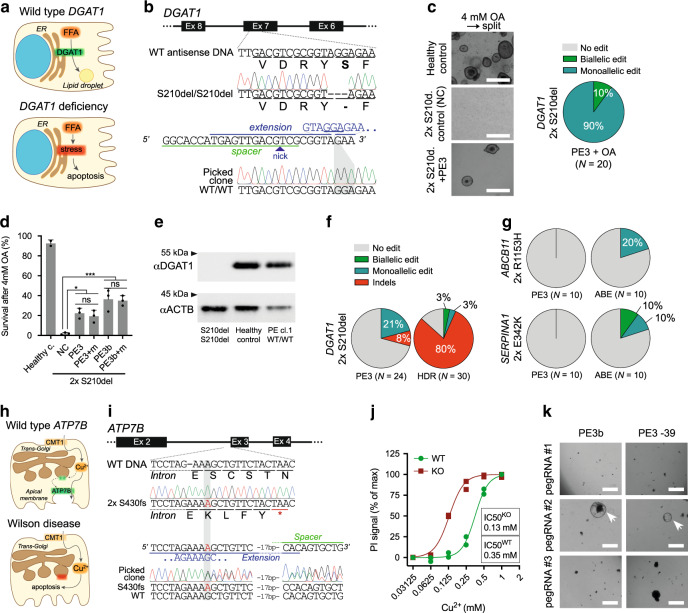
Fig. 2. Prime editing functionally corrects disease-causing indel mutations in intestinal and liver organoids.
a Schematic overview of the DGAT1 disease mechanism. b Sanger validation of biallelic DGAT1S210del mutations in patient-derived intestinal organoids, pegRNA design (nicking sgRNA at position +46 not shown), and Sanger validation of successful biallelic correction by PE3. c Brightfield images of healthy control- and DGAT1S210del patient-derived intestinal organoids (±PE3) after exposure to 4 mM OA for 24 h and subsequent passaging (split). White scale bars are 500 µm. Quantification of corrected alleles in patient organoids after PE3 and OA selection; all surviving organoids are gene-corrected. d Quantification of DGAT1S210del patient organoid survival upon exposure to 4 mM OA, after targeting with different PE3 or PE3b plasmids. Data are represented as mean ±S.D. of three independent experiments in two different donors. e Western blot of DGAT1 in DGAT1S210del, healthy control, and PE3-corrected DGAT1S210del organoids. f, Quantification of correct edits and unwanted indels by PE3 and Cas9-initiated HDR in DGAT1S210del organoids. Note that no functional selection with OA was performed prior to quantification. g Comparison of PE3 and adenine base editing (ABEmax-NG) to correct the ABCB11R1153H and SERPINA1E342K mutations in liver organoids from patients with BSEP-deficiency and alpha-1-antitrypsin deficiency, respectively. h Schematic overview of the Wilson disease (ATP7B deficiency) mechanism. i Sanger validation of biallelic ATP7BS430fs mutations in patient-derived liver organoids, pegRNA#2 design to target this mutation, and Sanger validation of successful monoallelic correction by PE3. j Cell death in ATP7BWT and ATP7BKO liver organoids after incubation with Cu2+ for 3 days. n = 2 biologically independent samples for both WT and KO groups. k Brightfield images of ATP7BS430fs-patient organoid survival upon exposure to 0.25 mM Cu2+ for 3 days, after transfection with different PE3 plasmids. “at −39” stands for nicking sgRNA at position −39. Note that only prime editing using pegRNA#2 yields functional correction of ATP7B (white arrowheads). White scale bars are 500 µm. ER endoplasmic reticulum, FFA free fatty acid, Ex exon, NC negative control, OA oleic acid, PE3+ m PE3 with introduction of PAM mutation, HDR homology-directed repair, ABE adenine base editor, PI propidium iodide. Source data are provided as a Source Data file.