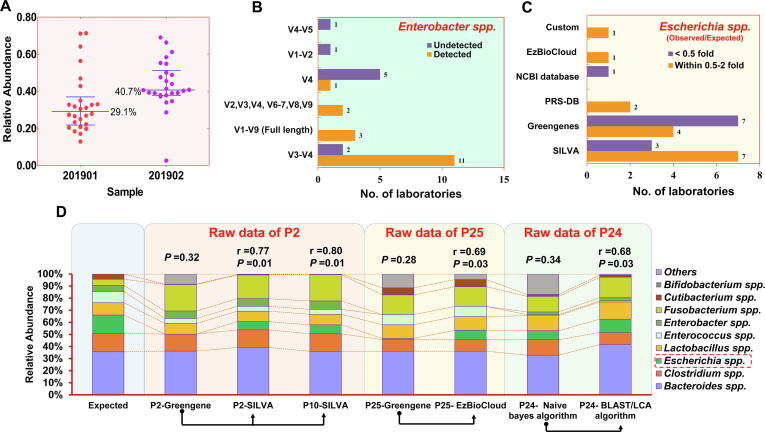
Fig. 4.
Potential sources of variation in 16Ss. (A) The cumulative relative abundance of G+ bacteria detected in the samples 201901 and 201902. Each point represents a participating laboratory. The line displays the interquartile range (lower quartile to upper quartile). The performance of the 16Ss laboratories in detecting Enterobacter spp. using different amplification regions (B) and in identifying Enterobacter spp. using different reference databases (C). (D) More accurate results were obtained by reanalyzing the raw data of the sample 201901 reported by several laboratories (P2, P25 and P24). The raw data of P2 were reanalyzed by P2 and P10 with pipelines including the SILVA database. The raw data of P25 were reanalyzed using the EzBioCloud database. The raw data of P24 were reanalyzed by changing the annotation algorithm. The correlations between the observed microbial abundances and the expected microbial abundances were calculated, and the corresponding Spearman's R and P values are shown above the histograms.