Significance
Information theory was invented to solve the task of sending reliable communication over an unreliable channel. T cells have extremely reliable signal-processing capacity as they sensitively and specifically respond to a few pathogen-derived peptide ligands displayed in the noisy environment of many self-derived ligands presented on the same antigen-presenting cell. We used information-theoretic concepts to analyze a computational model of the biochemical steps in the T cell signaling network to understand how features of the T cell signaling pathway enable discriminatory ability. Our calculations and experiments suggest that T cells superimpose kinetic proofreading steps that must be spatially localized with the receptor and feedback loops to extract reliable information from a noisy environment.
Keywords: T cell receptor ligand discrimination, self/agonist peptides, modeling, kinetic proofreading
Abstract
T cells exhibit remarkable sensitivity and selectivity in detecting and responding to agonist peptides (p) bound to MHC molecules in a sea of self pMHC molecules. Despite much work, understanding of the underlying mechanisms of distinguishing such ligands remains incomplete. Here, we quantify T cell discriminatory capacity using channel capacity, a direct measure of the signaling network’s ability to discriminate between antigen-presenting cells (APCs) displaying either self ligands or a mixture of self and agonist ligands. This metric shows how differences in information content between these two types of peptidomes are decoded by the topology and rates of kinetic proofreading signaling steps inside T cells. Using channel capacity, we constructed numerically substantiated hypotheses to explain the discriminatory role of a recently identified slow LAT Y132 phosphorylation step. Our results revealed that in addition to the number and kinetics of sequential signaling steps, a key determinant of discriminatory capability is spatial localization of a minimum number of these steps to the engaged TCR. Biochemical and imaging experiments support these findings. Our results also reveal the discriminatory role of early negative feedback and necessary amplification conferred by late positive feedback.
T lymphocytes (T cells) are critical in orchestrating an adaptive immune response to foreign pathogens and tumors. T cell receptors (TCRs) expressed on the surface of T cells recognize short peptides (p) bound to major histocompatibility complexes (MHCs). Selected in the thymus to bind relatively weakly to pMHC molecules derived from the host proteome, naive T cells circulate continuously after maturation, scanning the surface of antigen-presenting cells (APCs). During this time, the interaction with self pMHC is essential to maintain T cell survival. Concurrently, mature T cells search for pMHC molecules with peptides derived from pathogenic or mutated self proteins. If the bond between a particular TCR and foreign pMHC molecule (agonist ligand) is sufficiently strong, full T cell activation occurs. T cell detection of agonists on APCs is remarkably sensitive and selective: 1 to 10 agonists with binding half-lives only three times longer than thousands of null or weakly binding ligands can lead to activation (1–5). Moreover, the half-lives of stimulatory TCR–pMHC interactions are not very long (6, 7). How the T cell signaling network achieves this precise activation without mounting a full response to self pMHC has been the subject of numerous studies (3, 5, 8–15). However, most studies have focused on discrimination between self and agonist ligands presented separately.
The TCR signaling network discriminates between self vs. agonist pMHC (i.e., two possible inputs) to a degree that far exceeds what would be predicted by equilibrium considerations. Assuming equivalent on rates of binding for both ligands, the ratio of equilibrium association constants for TCR binding to agonist and self ligands is equivalent to the ratio of half-lives. The equilibrium constant is defined as the following:
| [1] |
where the quantities in brackets represent the concentrations of the pMHC species. If for agonist pMHC is three times higher than that for self, a threefold higher concentration of self ligands should generate the same number of bound TCR–pMHC complexes as agonist ligands. However, T cell discrimination remains robust for self ligand concentrations over many orders of magnitude. Therefore, equilibrium arguments cannot describe the observed ability of T cells to discriminate between APCs bearing a few agonist ligands in a sea of self ligands and APCs that only express self ligands.
Hopfield (16) and Ninio (17) proposed the nonequilibrium, kinetic proofreading model to explain the ability of biological systems to discriminate between two inputs with far higher selectivity than what equilibrium considerations (Eq. 1) would suggest. McKeithan (18) adapted the model to explain the ability of T cells to discriminate between self and agonist pMHC ligands. As per the kinetic proofreading model, the discriminatory capability depends on a single parameter: the lifetime of TCR–pMHC bonds, τ. Upon TCR–pMHC binding, a series of sequential intracellular biochemical steps must proceed before productive downstream signaling sufficient for activation ensues (3). Ligands that bind with a weaker half-life are more likely to unbind before these “proofreading steps” are completed, ending signal propagation. Most models regarding T cell sensitivity and selectivity rely on differences in half-life of TCR binding between nonstimulatory and agonist pMHC (11, 18–21). Recently, by employing optogenetic technology using blue-light sensitive protein LOV2 on a supported lipid bilayer and chimeric antigen receptors, Tischer and Weiner (22) were able to tune the half-life of receptor-ligand bonds by modulating the blue light intensity. For fixed receptor occupancy and load, their data showed that half-life was the strongest determinant of the downstream signaling response leading to diacylglycerol (DAG) accumulation. Concurrent optogenetic approaches from the Schamel laboratory (23) using the plant photoreceptor phytochrome B showed similar results for intracellular calcium increases. Both the increase in DAG and calcium following TCR stimulation result from the activation of phospholipase C (PLC). These are key biochemical events necessary to initiate T cell activation (24–27). Also, our recent work revealed that slow phosphorylation of a specific tyrosine in the adaptor LAT, known to recruit and promote PLCγ1 function, was important for the T cell to achieve ligand discrimination (28).
Three well-established conditions must be met to achieve error correction via the kinetic proofreading model. First, whether the bound ligand is self or nonself pMHC determines how far downstream the sequential biochemical reactions in the kinetic proofreading pathway progress. Agonist ligands that bind TCR with a longer half-life are more likely to be able to stay bound (“wait”) until the proofreading steps are completed. This implies that ligand release must trigger the disruption of progress along the proofreading steps. Second, the proofreading steps must be coupled to ATP consuming reactions that drive the system out of equilibrium. Only in a nonequilibrium system is there an arrow of time (difference between past and future), which provides meaning to the concept of a “waiting time.” Third, the net forward rates of the phosphorylation reactions cannot be too high; otherwise, signaling would quickly propagate down the sequence of steps even for self ligands that bind with a short half-life, and discrimination would not be selective.
Here, we used channel capacity to quantify how a biochemically faithful model of the T cell network uses kinetic proofreading to reliably extract information about the presence of a small number of agonist ligands in a noisy environment of many self ligands. Furthermore, our model seeks to elucidate how slow phosphorylation of Y132 on LAT may improve ligand discrimination. Our experiments strongly suggest that LAT phosphorylation is likely one of the minimum number of biochemical steps that must be spatially localized to the TCR after pMHC binding to achieve ligand discrimination. Finally, we describe how feedback loops superimposed on the kinetic proofreading steps enable the T cell signaling network to be an essentially noiseless communication channel. Negative feedback loops located close to the TCR suppress propagation of signals from TCR binding to self pMHC. Positive feedback loops located after ligand discrimination is complete results in digital amplification of signals from agonist ligands, thus enabling productive downstream signaling and functional discrimination.
Results
Channel Capacity Is a Direct Measure of T Cell Discrimination Capacity.
While the Hopfield error (16) is useful for quantifying the relative amount of erroneous output, it does not provide a direct measure of the T cell’s discriminatory capability. Consequently, analyses via information theoretic measures of ligand discrimination have been pursued (29–32). Lalanne and Francois (31) calculated the mutual information between ligand binding half-lives ( and ) and downstream biochemical signaling product. In reality, the T cell does not discriminate between two ligand types presented separately in equal concentrations on different APCs. Typically, it must distinguish between normal APCs displaying only self ligands and infected APCs that present at least one agonist ligand while also presenting many self ligands. Therefore, we cast the problem as one where the T cell signaling network must discriminate between two inputs: infected APCs that present a small number of agonist ligands (a bona fide signal) in a sea of self ligands and normal APCs that only display self ligands (tonic signals). If denotes the density of agonist ligands:
| [2] |
| [3] |
where denotes a fixed density of agonist ligands that is typically smaller than the background density of self ligands. Our studies are carried out over a distribution of the number of self ligands that may be presented on APCs. To quantify the extent to which the T cell signaling network can discriminate between normal and infected APCs, we use channel capacity, C, which measures a signal processing network’s ability to discriminate among inputs (33). In short, 2C quantifies the number of possible inputs that the network can discriminate: C = 1 means that perfect discrimination between two inputs is possible, and C = 0 means that the inputs cannot be discriminated at all.
Following the procedure detailed in SI Appendix: Derivation of Channel Capacity and Assumptions, which includes averaging over the log-normal distribution of self ligand densities (1, 34) on APCs, the channel capacity is defined mathematically as follows:
| [4] |
where is the mutual information of and intracellular biochemical output of TCR signaling that results in T cell activation. is the conditional probability distribution of the output given that the input APC either has no agonist ligands or a small number of agonist ligands , and .
There are two principal advantages of using channel capacity to quantify T cell discrimination: 1) Capacity allows quantification of discriminatory ability with explicit consideration of the variation in self ligand densities across APCs under normal conditions or during infection. This variation is reflected as the variance of the probability of obtaining a downstream signaling output given a value of Lf, . Channel capacity measures the extent of separation between this conditional probability distribution when the input is a normal vs. an infected APC (Fig. 1A). If the distributions overlap, a T cell cannot determine whether the APC is normal or infected based on intracellular output since there is a nonzero probability that it could be either. 2) Capacity has a clear upper bound: If C = 1, perfect discrimination of the inputs is achieved and any additional biochemical proofreading steps are unnecessary. Thus, channel capacity can quantify how the topology of the T cell signaling network accumulates information about the possible state of the input APC, thereby providing a direct measure of information transmission through the signaling network. Other metrics fall short of directly quantifying a T cell’s discrimination capacity in this manner (34–37).
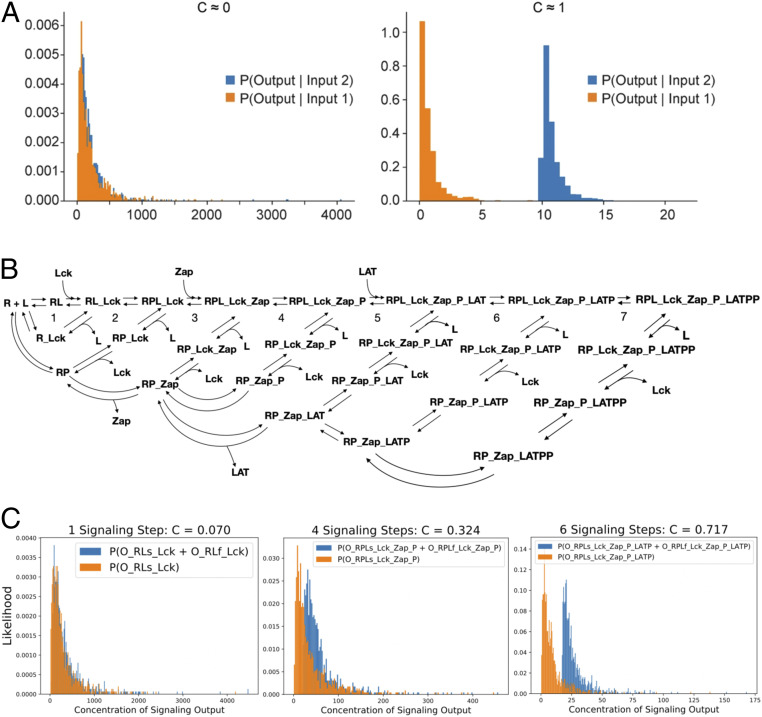
Fig. 1.
The discriminatory capacity of a realistic biochemical network describing the early events of TCR signaling in the context of kinetic proofreading. (A) Channel capacity (C) directly quantifies discrimination between inputs (e.g., normal and infected APCs). In the Left panel, output probability distributions from the two different inputs almost completely overlap, meaning that determination of input signal based on concentration of downstream output is not possible. On the Right panel, output distributions are disparate: If [Output] > 9 molecules, it is clear that input 2 has been sent; if less, input 1 was sent. (B) We consider two possible topologies of the TCR signaling network (see SI Appendix, Fig. S1 for species nomenclature): LAT remains associated with the TCR complex after being fully phosphorylated (RPL_Lck_Zap_P_LATPP) or, as depicted in SI Appendix, Fig. S3A, LAT dissociates from the complex (LATPP). (C) As kinetic proofreading steps are taken, the distribution of signaling output produced by cells that contain agonist and self peptides [blue, ] separates from the distribution of cells that contain only self [orange, ]. As a consequence, discrimination (channel capacity) increases as a function of the number of signaling steps taken.
Channel Capacity Increases with the Number of Kinetic Proofreading Steps.
The McKeithan model (18) unrealistically represents T cell signaling because direct dissociation of downstream signaling complexes to TCR and ligand do not occur. Also, biochemical steps along the main path are not completely irreversible. Instead, the TCR complex cycles through intermediate states back using the side paths, each cycle undoing part of the biochemical complex and modification (11, 14, 34, 38, 39). We studied a coarse-grained version of the early events in T cell activation where the most significant phosphorylation steps are included (Fig. 1B). Receptor (R) binds to ligand (L) leading to recruitment of CD4 or CD8 coreceptor-associated kinase Lck (40) to the TCR–pMHC complex (RL_Lck). Lck then phosphorylates the paired tyrosine residues of the immunoreceptor tyrosine-based activation motifs (ITAMs) on the ζ-chain or the CD3 chains associated with the engaged TCR (RPL_Lck), which is modeled as one step. Following ITAM phosphorylation, the tandem SH2 domains of ZAP-70 form a high-affinity complex with the doubly phosphorylated tyrosine sites in each ITAM (RPL_Lck_Zap) (41). Lck then phosphorylates residues Tyr-315 and Tyr-319 in the SH2-kinase interdomain B and Tyr-493 in the activation loop of ZAP-70 to generate catalytically active ZAP-70 (RPL_Lck_Zap_P). The SH2 domain of Lck binds phospho-Tyr-319 keeping ZAP-70 open and active as well as keeping Lck in close proximity and promoting its open, active conformation (42). After activation of ZAP-70’s catalytic activity, LAT is phosphorylated by ZAP-70 at multiple sites (RPL_Lck_Zap_P_LATP and RPL_Lck_Zap_P_LATPP) (43). For reference, SI Appendix, Fig. S1 defines nomenclature of all pertinent species.
Reversal of biochemical modifications is triggered by unbinding of ligands. As an example, in cycle 3, ligand dissociation (from RPL_Lck_Zap to form RP_Lck_Zap) causes coreceptor associated Lck to leave first. ZAP-70 protects phosphorylated ITAMs from de-modification until it unbinds and allows phosphatases to de-phosphorylate the tyrosine residues and form free receptor. Similar arguments apply to other cycles, and many more possible side paths exist than those illustrated in Fig. 1B. Assuming rapid disassembly of the TCR complex occurs after ligand dissociation, we do not anticipate qualitative differences with regards to discrimination if different permutations are explored. The ordinary differential equations describing the reaction networks in Fig. 1B were solved numerically via the PySB software package (44). The “Real-valued Variable- coefficient Ordinary Differential Equation solver” (VODE) with fixed-leading-coefficient implementation was used to integrate the equations (45).
Fig. 1C shows how the channel capacity, and concomitantly, the T cell’s ability to discriminate between normal and infected APCs, increases as additional proofreading steps are completed. After Lck recruitment, the conditional output distributions for cases with (blue) and without (orange) agonist ligands overlap completely such that C ∼ 0.07 and discrimination is not possible. After each step in the signaling pathway, the distributions separate further and capacity increases until complete separation and perfect discrimination is reached after full phosphorylation of LAT (given ).
Assuming that the free concentrations of signaling proteins are in excess, the rate of change in channel capacity depends on the rate constants along the signaling path and the variance of the self ligand distributions. If self ligands are log-normally distributed with a larger variance, there will be greater overlap in the conditional output distributions and either more proofreading steps or slower phosphorylation rates will be necessary to achieve the same capacity increase per step. As explored later in the text, the change in capacity depends most significantly on .
Kinetic proofreading increases channel capacity for two reasons. First, signaling steps are non-Markovian; that is, they are not independent of past and future steps. T cell signaling would be a Markov process if the steps proceeded as follows: , where and are downstream signaling products formed at steps and . For such a Markov process, the data processing inequality (33) dictates that , i.e., information about the ligand contained after steps (in ) must exceed that after steps (in ). As an analogy, if a person acquires information from a source and passes it on to a second person, who then tells a third person, etc., then the third person cannot have more information from the source than the second person as some information is likely lost during successive tellings of the story. TCR signaling (Fig. 1B) is not a Markov process because the rates of side paths depend upon whether self or agonist pMHC molecules bind to the TCR in the first signaling step; i.e., the occurrence of a step depends on past history. Thus, violation of the data processing inequality is possible. Next, biochemical steps along the main path must be driven out of equilibrium, which is the case in Fig. 1B since most of the modifications are tyrosine phosphorylation reactions that hydrolyze ATP and dissipate energy. If these steps proceeded at equilibrium, information gained by taking a forward step would be lost by an equally likely reverse step.
Speed of Phosphorylation of LAT Residues Affects Channel Capacity and Discrimination Capability.
LAT has five key tyrosine phosphorylation sites, with Tyr-132 being the slowest step due to a preceding glycine residue (G131) (46–48). Recently, we (28) showed that substituting glycine with a negatively charged glutamate or aspartate accelerates Tyr-132 phosphorylation and endows T cells with the ability to respond to APCs presenting only weak and even self pMHCs. In information-theoretic terms, accelerating Tyr-132 phosphorylation causes a drop in channel capacity. We explored the discriminatory effects of slow Tyr-132 phosphorylation using our computational model and determined the possible mechanisms underlying the decrease in capacity.
Since our primary focus is to understand how network topology and relative rates of LAT phosphorylation steps affect T cell discrimination, exact numerical values of the rate constants are less significant. All reaction rates and initial concentrations of signaling products are described in SI Appendix, Rate Constants for Simulations. Forward rates for the biochemical steps on the main path cannot be too fast or discrimination will not be possible; if rates are too slow, perfect discrimination will be reached in only a few proofreading steps such that the signal strength would become too small to be propagated by the time LAT is fully phosphorylated. Given the observed importance of Tyr-132 phosphorylation on ligand discrimination (28), we chose intermediate values of the forward rates: and 2.0 s−1 depending on network topology.
Currently, it is unknown whether Tyr-132 phosphorylation is a kinetic proofreading step or whether LAT dissociates from the TCR complex after full phosphorylation (43, 49, 50). We explored the effects on capacity in both cases. In Fig. 1B, the first four LAT phosphorylation steps are treated as a single step, resulting in production of RPL_Lck_Zap_P_LATP. Clearly, if LAT remains attached to the TCR complex even after full phosphorylation, Tyr-132 phosphorylation is a non-Markovian kinetic proofreading step. If C < 1 before Tyr-132 phosphorylation happens, Fig. 2A shows that slowing the step down (, red line) increases channel capacity and enhances APC discrimination. The step may be especially slow because it is the final opportunity for the T cell to reach perfect discrimination before downstream signaling products dissociate from the TCR complex. After dissociation, the ensuing biochemical steps are Markovian and channel capacity can no longer increase, in accordance with the data processing inequality. In our model, slowing down an earlier step (SI Appendix, Fig. S2A, gray) results in the same increase of capacity. This is because the lifetime of signaling complexes remained constant after each step along the main path. In reality, due to accumulation of bound species, signaling complexes become more stable as steps proceed, making slower phosphorylation of later steps more critical to achieve proofreading.
Fig. 2.
Slow LAT Y132 phosphorylation and spatial localization. (A) depicts capacity vs. number of signaling steps for the T cell network presented in Fig. 1B where Y132 phosphorylation is a kinetic proofreading step: Speeding up Y132 phosphorylation via G131 mutation yields the green curve, and wild type (WT) is given by the red curve. The corresponding curves for the network shown in SI Appendix, Fig. S3A are the blue (G131 mutation) and orange (WT) curves; the Inset shows the drop in capacity due to the G131 mutation. (B) Numerical calculations of (minimum number of ligand-associated steps) derived from the network shown in SI Appendix, Fig. S4: For a fixed set of rate constants (SI Appendix, Table S4), is calculated by finding the smallest value of(number of ligand-associated steps) that gives an error rate . The calculation is repeated for a range of (ratio of half-lives of agonist to self ligands) and two values of (ratio of concentrations of agonist to self ligands).
Alternatively, as explored in SI Appendix, Fig. S3A, LAT may dissociate from the TCR complex after full phosphorylation. In this case, Tyr-132 phosphorylation is a Markovian step and the data processing inequality dictates that channel capacity can either remain constant or decrease. Surprisingly, if Tyr-132 phosphorylation proceeds at the same rate as the preceding step (Fig. 2 A, Inset, blue line), we observed a drop in discrimination. The drop is more pronounced for faster on-rates (SI Appendix, Fig. S3B). The histograms in SI Appendix, Fig. S3C show that the capacity drop can be attributed to the excess catalysis of downstream signaling product (LATPP) by APCs that contain high self-pMHC densities (see red box, orange histogram). Corresponding to the kinetics observed in wild-type LAT, slowing Tyr-132 phosphorylation prevents “leakage” of these partially phosphorylated LAT molecules from propagating further (SI Appendix, Fig. S3C, red box) causing capacity to improve to its previous value (Fig. 2 A, Inset, orange line) and averting a drop in discrimination.
Our computational results mirror the experimental results: Slow phosphorylation of Tyr-132 improves ligand discrimination whether or not fully phosphorylated LAT remains attached to the receptor complex, but for different reasons. If LAT remains localized with the receptor even after full phosphorylation, the underlying mechanism relies on Tyr-132 phosphorylation being a bona fide kinetic proofreading step.
Signaling Components Must Remain Spatially Proximal to the TCR for a Minimum Number of Steps for Proper Ligand Discrimination to Occur.
The preceding calculations suggest that slowing Tyr-132 phosphorylation on LAT would improve discrimination between normal and infected APCs if fully phosphorylated LAT was spatially colocalized with the TCR–pMHC complex. Given that spatial proximity of LAT to the TCR is critical for achieving high capacity, we explored the possibility that there is a minimum number of biochemical steps that follow TCR–pMHC binding , which must also remain spatially localized to the receptor complex. should depend primarily on , the ratio of binding half-lives of agonist and self ligands, as well as , the ratio of agonist to self ligand densities on infected APCs. Using a simplified kinetic proofreading model (SI Appendix, Fig. S4), we semianalytically derived the dependence of on these variables.
In this model, after a fixed number of biochemical steps, a product denoted by loses knowledge about the ligand after it dissociates from the molecular complex that remains connected to the receptor. then acts as an enzyme and phosphorylates a downstream substrate to produce . Solving the differential equations describing the network at steady state while assuming that gives the following formula for the Hopfield error rate (SI Appendix: Toy Model):
| [5] |
where , , is the phosphorylation rate, is the off-rate of self ligands, and is the off-rate of agonists. The steady-state values of and are given by SI Appendix, Eqs. S19 and S21.
We picked values (SI Appendix: Toy Model) for parameters and found necessary to reach (SI Appendix, Fig. S5), an error rate that corresponds to the approximate upper bound on T cell discriminatory accuracy (18). Fig. 2B shows that decreased as and increased, with a much stronger dependence on the ratio of binding half-lives. In other words, if self and agonist ligands have more similar half-lives and if the concentration of self ligands is much greater than that of agonists on the surface of APCs, more signaling steps must occur near the TCR—results that make intuitive sense.
Since does not directly measure a T cell’s capacity to discriminate between normal and infected APCs, we derived an expression for the minimum number of spatially localized signaling steps necessary before , (SI Appendix: Toy Model). With the same set of parameters, we calculated (SI Appendix, Fig. S6) and observed a dependence on (SI Appendix, Fig. S7) that is consistent with what was found in Fig. 2B.
Experimental Tests Show that LAT Is Colocalized with the Receptor, and This Is Important for Ligand Discrimination.
The computational results via the information-theoretic metric of channel capacity revealed how various features of the T cell signaling network enable ligand discrimination. These results suggest that the experimental observation of slow Tyr-132 phosphorylation on LAT is important for ligand discrimination could be either because this is an important kinetic proofreading step, or not. LAT phosphorylation is likely among the necessary minimum biochemical steps colocalized to the TCR, and thus, disruption of colocalization of LAT to the TCR can diminish discrimination ability. We have tested these predictions using single-molecule imaging and biochemical experiments.
Single-molecule pMHC binding events result in LAT assemblies spatially localized to the receptor–ligand complex.
Single-molecule imaging of mouse CD4 T cells expressing the AND transgenic TCR, which recognizes a moth cytochrome c (MCC88–103) peptide, revealed LAT’s colocalization with agonist pMHC-engaged single TCR complexes (Fig. 3). The spatial localization of downstream signaling events initiated by pMHC:TCR binding was assessed by total internal reflection fluorescence microscopy. Imaging was performed on transgenic AND TCR T cells interacting with functionalized supported membranes containing ICAM-1 (∼600 molecules/μm2) and the AND TCR agonist ligand, MCC peptide loaded onto MHC. The peptide is synthesized with an Atto647N fluorescent label, loaded onto MHC class II molecules, and presented at low density (0.07 molecules/μm2) on the supported membrane. At this ligand density, full T cell activation (as measured by NFAT translocation) is not generally achieved, but the early signaling activity from individual pMHC:TCR complexes is readily resolved.
Fig. 3.
Single-molecule pMHC binding events produce localized LAT assemblies: (A) Fluorescently labeled pMHC were adhered to a supported lipid bilayer, along with the adhesion molecule ICAM-1. T cells landing on the bilayer were imaged for 180 s. (Left) Green reticles indicate slow-moving (bright) pMHC:TCR complexes. (Middle) Red reticles indicate significant LAT assemblies. (Right) Overlay of pMHC and LAT indicated colocalization. (B) Images of 14 separate binding events that occurred during the acquisition of the cell featured in A. The time point shown for each binding event was when the localized LAT assembly reached its maximal size. (C) Bar plot of initial center–center distances of centroids detected over pMHC and the LAT assembly. (D) Intensity histogram of pMHC-Atto647 point spread functions fit to a log-normal distribution. (E) Trace of single-molecule binding showing single-step bleaching and Poisson noise. (F) Intensity distribution of individual LAT-eGFP molecules within the cell after bleaching down to single molecules fit to a log-normal distribution. The mean of this distribution along with a Western blot was used to estimate the total number of LAT molecules within the clusters.
At long exposure times (500 ms) (51) and low ligand densities, individual slow moving pMHC:TCR complexes are distinguishable from free unbound pMHC. Fig. 3A is an image of such pMHC:TCR complexes, which are identified by green reticles. Primary AND T cells, retrovirally transduced with LAT-eGFP, were added to the imaging chamber, allowed to adhere onto the ICAM/pMHC supported membrane, and imaged for 3 min. Both the Atto647N-MCC-MHC and LAT-eGFP channels were resolved simultaneously with a dual-camera setup. Long dwelling pMHC:TCR binding events (20 to 50 s) frequently resulted in spatially localized LAT assemblies (Fig. 3 A and B) that began forming within 50 to 80 nm of the ligated TCR (Fig. 3C). The distances calculated are from the center of the ligating pMHC to the center of initial LAT clustering. Typical localization error for superresolution experiments (with stationary fluorophores) is around 20 to 30 nm. In our experiments, the bound pMHC move slowly (∼0.1 μm/s) over the 0.5-s exposure, adding ∼50 nm of localization error. That the observed deviation between pMHC and LAT (50 to 80 nm) paralleled the fundamental localization error (50 to 70 nm) suggests that they are strongly colocalized.
After initiation, some assemblies drifted a small distance (∼300 nm) away from the receptor. Characterization of the Atto647N-pMHC on the bilayer showed them to be isolated monomers (Fig. 3 D and E), whereas the LAT assemblies produced contained hundreds of LAT molecules (Fig. 3F). These results indicate that LAT phosphorylation and condensation, via the LAT:Grb2:SOS protein binding network (52–54), is highly localized with the pMHC:TCR binding events.
Experiments interfering with LAT’s association to the TCR complex show decline in ligand discrimination capability.
Next, we experimentally tested whether spatial localization of LAT to the TCR–pMHC complex is important for discrimination, and thus, whether LAT-involved biochemical events are necessary proofreading steps. Toward this end, we utilized Jurkat cells, a human lymphoblastic T cell line. As reported previously, OT-I+ hCD8+ J.Lck.Lck-FLAG Jurkat mutant (J.OT-I+) was able to use the OT-1 TCR to recognize the agonist OVA peptide, which is derived from the chicken ovalbumin protein (OVA257–264) when presented by mouse MHC class I molecules H-2Kb (43). An OVA altered peptide ligand, T4, is a weaker agonist with affinity at the threshold just strong enough to promote thymocyte negative selection (55). To examine how LAT proximity to the engaged TCR complexes affects discrimination capability, we overexpressed a CFP-tagged LAT mutant in which all nine tyrosine residues were substituted with phenylalanine (LAT-CFP-Y>F) to abrogate its ability to promote TCR signal transduction (47, 56). We compared the impact of overexpression of “all-F” mutants that either harbored a functional proline-rich PIPRSP motif or did not (LAT-CFP-Y>F-PIPRSP or LAT-CFP-Y>F-AIARSA, respectively). Since, as we previously described (43), the PIPRSP motif can facilitate LAT’s association with the engaged TCR complexes, by interacting with the Lck SH3 domain contained therein, mutations of proline to alanine in this motif would decrease the “all-F” LAT’s ability to remain close to the TCR. Thus, “all-F” LAT harboring the proline mutations does not compete with the ability of endogenous LAT to be recruited to the engaged TCR complexes. On the other hand, overexpression of the “all-F” LAT with a functional PIPRSP motif can associate with the engaged TCR complexes, thus acting to decrease the recruitment of endogenous LAT to the engaged TCR complexes.
If the two types of LAT mutants are overexpressed respectively, any differences in signal propagation should reflect their relative abilities to compete with endogenous LAT to be recruited to the engaged TCR complexes (Fig. 4A). In this experimental setting, if the proximity of LAT to the engaged TCR complexes has no impact on TCR signal efficiency, we should not observe differences in the ligand discrimination responses between cells overexpressing LAT-CFP-Y>F-PIPRSP or LAT-CFP-Y>F-AIARSA. However, upon examining calcium responses (Fig. 4B) stimulated by either the full agonist OVA/H-2k or partial agonist T4/H-2k, we found this was not the case. During the initial phase of stimulation, J.OT-I+ cells expressing LAT-CFP-Y>F-AIARSA (a noncompetitive antagonist; referred to as AAA hereafter) showed stronger calcium influx compared with cells expressing LAT-CFP-Y>F-PIPRSP (a competitive antagonist; referred to as PPP hereafter). The Left panel of Fig. 4B shows that when individual engineered cells were stimulated with OVA (at 30 s when SA was added), both PPP and AAA responded quickly to comparable degrees, as evidenced by the observation that the Indo-1 ratio increased at ∼35 to 40 s immediately after the addition of OVA stimuli (gray shadow). In contrast, when the T4 stimulus was added (Fig. 4 B, Right), we observed a smaller increase in the Indo-1 ratio during the initial phase and a much smaller response for PPP. These results demonstrate that LAT-CFP-Y>F-AIARSA (AAA) interfered less with the recruitment of endogenous LAT to the engaged TCR complexes because it lacks the functional PIPRSP motif.
Fig. 4.
The recruitment of LAT to the proximity of engaged TCR complex enhances signal efficiency: (A) The representative dot plot (Left) shows the relative ectopic expression level of CFP-tagged LAT Y>F mutant. The cells expressing AIARSA or PIPRSP LAT-CFP-Y>F were labeled with different ratios of Cell Trace Yellow dyes, allowing us to pool the cells in one tube for the following stimulation and calcium experiments. The cartoon (Right) illustrates the experimental design. (B) The representative calcium flux plot for cells stimulated with OVA/H-2Kb (Left) or T4/H-2Kb (Right). Cells were labeled with calcium-sensitive dyes Indo-I, washed twice, and then labeled with OVA/H-2Kb or T4/H-2Kb biotinylated monomers at 37 °C for 5 min and analyzed on a BD LSRFortessa for calcium analyses. Cells were first recorded for 30 s to establish the baseline, and the streptavidin (SA) was added to stimulate the cells. (C) The representative bar plot of the Indo-I ratio normalized with respect to baseline during the initial phase of stimulation with OVA/H-2Kb and T4/H-2Kb. (Left) The red bars quantify the normalized ratios when high concentrations of LAT-DN-AAA (Control) are added to the cell, and the blue shows the response when high LAT-DN-PPP is added. (Right) The change in the normalized response due to interference by high LAT-DN-PPP.
Upon closer examination, we find that interference with localization of LAT to the TCR complex played an even greater role for discriminating weak ligands from self. In the Left panel of Fig. 4C, we quantify the differences in responses for OVA and T4. Normalizing the Indo-1 ratio in the initial phase with respect to the baseline response, which represents the “no activation” scenario similar to the condition of self-antigen stimulation, the red bars show that discrimination with respect to self is significantly better for OVA (strong agonist) than for T4 (weak agonist) stimulation. Interference with LAT association to the TCR (High LAT-CFP-Y>F-PPP) leads to a drop in discrimination with respect to self for both OVA and T4 as shown by the blue bars. Importantly, the drop in T4 response is so large that discrimination with respect to self becomes no longer possible. These results show that spatial localization of LAT to the signaling complex vicinal to the TCR is critical for signal discrimination.
The Right panel (Fig. 4C) plots the difference between the red and blue bars shown in the Left panel (Fig. 4C) for OVA and T4. Crucially, the drop in discrimination for T4 is greater than that for OVA. The result is consistent with the predictions shown in Fig. 2B: A minimum number of signaling steps must remain associated with the receptor to achieve proper discrimination between self and agonist ligands. increases as the ratio of half-lives of agonist to self-ligands decreases. So, T4 is predicted to require a greater number of localized signaling steps compared to OVA. The experiments show that LAT association with the TCR is likely one of those minimum steps for T4: T4 better represents most foreign antigens, as OVA is an unusually strong agonist. Given the discriminatory role of slow LAT phosphorylation (28) (Fig. 2A) and strong evidence in support of the Lck: LAT “bridge” model (43), we conclude that LAT association with the TCR is a critical part of the kinetic proofreading cascade that must be spatially localized with the receptor.
Feedback Regulation of Kinetic Proofreading in the T Cell Signaling Network.
Our results thus far have clarified how the earliest biochemical steps in T cell signaling mediate kinetic proofreading, and importantly, that biochemical events occurring at LAT are part of the kinetic proofreading steps that must be spatially localized with the receptor complex. However, the early steps in the TCR signaling network also contain feedback loops. For example, a positive feedback loop downstream of LAT that is initiated by SOS-mediated Ras activation is well established (57–60). More recently, a negative feedback loop involving activated ZAP-70 inhibiting Lck activity has also been reported (61, 62). The role of feedback regulation has also been studied in the context of kinetic proofreading and T cell discrimination (34–36), with models that emphasize the competing roles of SHP-1 (negative) and ERK (positive) feedback. However, recent mouse studies do not support these models (63). To our knowledge, none of the past models explicitly tested how feedback loops regulate T cell discrimination of normal and infected APCs, an issue we study using our metric of channel capacity.
Early negative feedback enhances ligand discrimination and sensitivity to agonists.
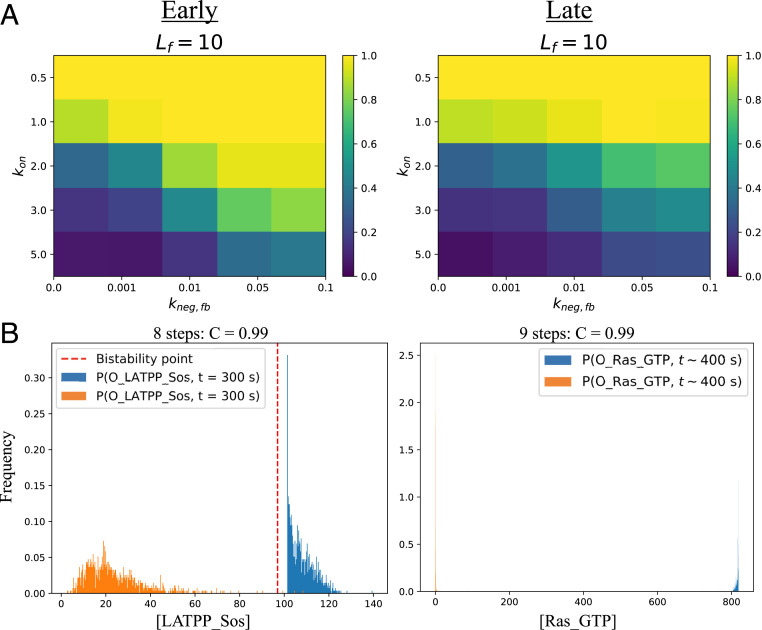
We first examined the effects of ZAP-70 inhibition of Lck on channel capacity by adding the biochemical reactions shown in SI Appendix, Fig. S8A to those depicted in SI Appendix, Fig. S3A. Phosphorylated ZAP-70 due to binding of both self and agonist pMHCs catalyzes the conversion of active Lck into an inactive form . For a range of forward rates and feedback strengths , we computed the channel capacity and found that negative feedback improved T cell discrimination (Fig. 5A, Early). Here, negative feedback acted as a bottleneck, slowing down early proofreading steps through Lck inactivation. Since most APCs contain self ligands in vast excess over agonist ligands, early negative feedback overwhelmingly inhibits signals from self ligands especially in APCs that present them in high concentrations.
Fig. 5.
The role of feedback loops: (A) comparison of early and late negative feedback. Color bar indicates value of capacity measured in bits (yellow, C ∼ 1; blue, C ∼ 0). In the early case, ZAP-70 bound to the TCR complex inactivates the free pool of active Lck. For late feedback, RPL_Lck_Zap_P_LATP inactivates the free pool of ZAP-70. Early negative feedback is more effective at improving discrimination. (B) Ras-SOS positive-feedback loop. If parameters are chosen (SI Appendix: Positive Feedback Loops) so that RasGTP-bound SOS is more effective at catalyzing formation of RasGTP than RasGDP-bound SOS, there will be a bistable point. (Left) Discrimination is nearly perfect by the time LATPP recruits SOS. (Right) If the bistable point is located between the distributions, signal (blue) is amplified and external noise (orange) is quenched.
Given that signaling output from self ligands diminishes as proofreading steps are taken, we expect that the same type of negative feedback loop acting later would be less effective at improving ligand discrimination. To study the effect of late negative feedback, we supposed that after the formation of the transient complexes Lck/ZAP-70/LAT with the engaged TCR (RPL_Lck_Zap_P_LATP), ZAP-70 was inactivated and thereby prevented from attaching to ITAMs (SI Appendix, Fig. S10A). Fig. 5A shows that early negative feedback was more effective in improving discrimination than late feedback, a difference that became even more pronounced for faster kon. Additional computational results showed that perfect discrimination was achievable with a single agonist pMHC present if negative feedback happens early (SI Appendix, Fig. S10B), suggesting that it allows T cells to attain high sensitivity.
Late positive feedback digitally amplifies agonist signals to result in functional discrimination between normal and infected APCs.
As signal propagated down the TCR signaling pathway, discrimination improved due to kinetic proofreading, but signal strength (number of activated molecules within all T cells) decreased (Fig. 1C, see the scale of the abscissa). If TCR signal strength is too low, active molecules will be unable to send sufficiently strong downstream signals for new gene transcription programs required for T cell activation and there would be no functional response to agonists. The Ras-SOS pathway acts downstream of the initial TCR signaling steps: SOS (Son of Sevenless) is recruited to LAT and catalyzes the conversion of Ras-GDP to Ras-GTP (57). SOS’s catalytic activity is enhanced when Ras-GTP binds to its noncatalytic, allosteric site, thereby creating a positive feedback loop (58–60). The biochemical reactions (SI Appendix, Fig. S11A) describing the Ras-SOS allosteric pathway leads to a bistable or digital dependence of Ras-GTP production on LATPP-SOS (SI Appendix, Fig. S11B) (8). It is possible that digital signaling induced by feedback regulation of SOS’s GEF activity, coupled to kinetic proofreading steps allows functional responses to infected but not normal APCs.
Indeed, we observe this effect by connecting the SOS feedback loop to our kinetic proofreading network (SI Appendix, Fig. S3A) while including early negative feedback of Lck (SI Appendix, Fig. S8A). Within the typical timescale of activation (1∼5 min), for kon = 1.5 s−1 (SI Appendix, Figs. S12A and S13A), after SOS recruitment, very little overlap remains between the output distributions of infected and normal cells (Fig. 5 B, Left). Assuming a bistability point at [LATPP-SOS]crit ∼ 97 (Fig. 5 B, Left, red dotted line), the RasGTP is digitally amplified when agonist ligands are present, and output from T cells interacting with APCs containing only self ligands is quenched (Fig. 5 B, Right). If the same phenomenon were to happen earlier, i.e., if RPL_Lck_Zap were to be subjected to positive feedback regulation (SI Appendix, Fig. S14), then output in the presence of self-ligand–only stimulation would be amplified, leading to a significant drop in discrimination (SI Appendix, Fig. S15). T cells must wait until APCs that present maximal concentrations of only self ligands (right tail of orange histogram in Fig. 5 B, Left) is less than output generated by APCs that contain minimal concentrations of agonist and self ligands (left tail of blue histogram in Fig. 5 B, Left). Therefore, information about typical surface ligand concentrations on different APCs may be encoded in the placement of the positive feedback loop.
Discussion
A proper T cell-mediated response relies on the ability to accurately and promptly react with a bona fide activating signal. However, the task is complicated by the fact that T cells also continually interact with self peptides to generate tonic signaling necessary for their survival. Thus, T cells have to properly distinguish between agonist pMHC stimuli in a noisy background of self pMHC. With the intense research interests and abundance of data regarding TCR proximal signaling, we now have a clearer understanding of how TCR signals are initiated, amplified, and propagated downstream (64). However, only recently have we begun to understand how proximal TCR signaling networks involve kinetic proofreading to ensure signal accuracy.
Here, we define a direct measure of T cell discrimination—viz. channel capacity, which allows us to appropriately account for the abundant distribution of self pMHC on APCs and determine how absolute ligand discrimination is achieved through kinetic proofreading. First, we computationally studied a reasonably realistic model for early TCR signaling events, with explicit account of the distribution of a large number of self ligands across APCs. Our results reveal how discrimination improves, i.e., channel capacity increases, because of the nonequilibrium and non-Markovian nature of the kinetic proofreading steps that reverse biochemical transformations upon ligand unbinding (33). The dependence of information entropy reduction (channel capacity increase) on thermodynamic entropy production due to ATP consumption in this model is consistent with the thermodynamics of information (65). Acquisition of information can drive a physical system out of equilibrium with no apparent energetic cost (65, 66). In this case, the T cell must expend an optimal amount of energy to gain information about the extracellular environment. If the phosphorylation rate proceeds too quickly, self-ligand output increases and capacity may decrease: What has already been gained may be lost (Fig. 2A). Our findings provide a number of possible explanations for the experimental observation that slow phosphorylation of Tyr-132 on LAT enhances discrimination capability. If Tyr-132 phosphorylation is a kinetic proofreading step (Fig. 1B), the slow step is necessary to increase channel capacity. As the TCR–pMHC complex accumulates bound species, i.e., Lck and ZAP-70 with each signaling step, it is possible that more time is needed for complex disassembly after unbinding of self ligands. If it does not proofread (SI Appendix, Fig. S3A), the slow step prevents a drop in capacity due to propagation of noise from APCs that express high concentrations of self ligands.
We also described several essential concepts: 1) There is a minimum number of kinetic proofreading steps, 2) these steps must be spatially localized with the receptor complex for the T cell to achieve proper discrimination between infected and normal APCs, and 3) the minimum number of steps is greater if the T cell has to discriminate between self and agonist ligands with more similar half-lives or if the T cells need to detect fewer agonist ligands on APCs. The calculations presented in Fig. 2B suggest that these concepts evolved in response to typical lifetime and concentration ratios of self and agonist ligands presented on APCs.
Our study supplements increasing evidence that spatial localization of Lck, ZAP-70, and LAT is critical for signaling (67–69). The SH2 domain of Lck binds to phospho-Tyr-319 in interdomain B of ZAP-70, sustaining Lck localization and activity while also stabilizing the active conformation of ZAP-70. Interestingly, Lck cannot phosphorylate LAT or SLP76, known substrates of ZAP-70 (48). Based on recent experiments, Lo et al., (43) proposed that the SH3 domain of Lck forms close contacts with the proline-rich PIPRSP motif in LAT; Lck’s interaction with ZAP-70 via its SH2 domain and its contact with LAT allows it to form a bridge between ZAP-70 and LAT. In doing so, sustained phosphorylation of LAT by active ZAP-70 is possible, allowing effective signal propagation (43). Our experimental data using a competitive antagonist construct of LAT validate the preceding hypothesis, and our computational modeling suggests that the number of biochemical steps that must be localized with the receptor includes LAT.
The “bridge” model is preferable to the “catch-and-release” mechanism proposed by Katz et al. (49), which suggests that ZAP-70 after activation by Lck dissociates from the TCR, translocates to spatially distinct nanoclusters of LAT, and activates them. The latter mechanism is difficult to reconcile with our results. If active ZAP-70 unbinds from the TCR–pMHC before steps are taken (SI Appendix, Fig. S16A), discrimination capacity drops with respect to the case where it remains bound (SI Appendix, Fig. S16B). If LAT is not spatially localized with the receptor complex by a mechanism akin to bridging, slow Tyr-132 phosphorylation of LAT would play no role in improving discrimination.
We investigated the role of regulatory feedback loops in affecting T cell discriminatory capacity. The results showed that to achieve high capacity, negative feedback loops must occur at earlier steps to prevent inappropriate initiation of signals by self pMHC. This behavior is observed since APCs are more likely to present self pMHCs in high concentrations relative to agonist pMHCs. One such negative feedback circuit could be represented by the ZAP-70–mediated phosphorylation of Tyr-192 in the Lck SH2 domain, which would negatively impact the ability of CD45 to positively regulate Lck (61, 62). There is some evidence that receptor-proximal negative feedback can quench “noise” to improve discrimination. Csk negatively regulates SFKs by phosphorylating their negative regulatory sites. As SFKs are activated, they phosphorylate a number of adaptors (i.e., PAG85), which can recruit Csk to the membrane. This places Csk in close proximity with SFKs to phosphorylate the C-terminal negative regulatory site in SFKs to dampen their activities. In Courtney et al. (70), we have shown that inhibition of Csk changes the threshold stimulation for weak but not strong agonists. Also, neither the sensitivity, nor the maximum response to stronger agonists change.
As kinetic proofreading steps proceed to improve T cell discrimination, the output of the biochemical reactions continues to become smaller. Our results suggest that positive feedback loops located further downstream can then amplify the signal strength digitally, converting them into significant downstream signals that lead to activation, and thus enabling T cells to functionally respond to infected, but not to normal, APCs. There is evidence that positive feedback loops distal from the receptor aid functional ligand discrimination. This is perhaps best exemplified by the action of RasGRP generated Ras to amplify SOS activation. In Das et al. (8), a small amount of DAG generated by PLCγ1 activation leads to RasGRP activity, which generates small amounts of Ras; the production of Ras can prime the allosteric site of SOS whose activity is promoted. An analog Ras signal generated by DAG increase leads to digital Ras pathway signaling by SOS activation—i.e., positive feedback postkinetic proofreading promotes functional discrimination. In the future, it will be important to explore the role of the recently studied LAT condensation process (53, 54, 71–74) on positive feedback regulation following LAT phosphorylation.
Our results suggest that a minimum number of kinetic proofreading steps spatially localized near the TCR–pMHC complex (that includes LAT phosphorylation), further augmented by early negative and late positive feedback regulation, enables the T cell signaling pathway to function as a noiseless communication channel that exhibits extraordinarily high sensitivity and specificity in detecting pathogen-derived TCR ligands.
Materials and Methods
Cell Lines.
The OT-I+ hCD8+ J.Lck.Lck-FLAG Jurkat mutant (J.OT-I+) was maintained in RPMI culture medium supplemented with 5% fetal bovine serum and 2 mM glutamine (43).
Calcium Experiments.
The J.OT-I+ cells were electroporated to overexpress pEF-vectors that consist CFP-tagged LAT mutant LAT-CFP-Y>F-PIPRSP, or LAT-CFP-Y>F-AIARSA. The CFP-tagged “all-F” LAT mutant was generated as previously described (47). The mutant AIARSA motif was introduced to the wild-type CFP-tagged “all-F” LAT construct by site-directed mutagenesis using QuikChange Lightning (Agilent). A total of 1 × 107 J.OT-I+ cells was electroporated with 10 μg of individual plasmids under the following conditions: 1,250 μF, 260 V, ∞ resistance using 0.4-cm Gene Pulser cuvettes (Bio-Rad). LAT-CFP-Y>F-PIPRSP cells were labeled with CellTrace Yellow dyes (Thermo Fisher Scientific). Then, cells were rinsed with PBS twice. LAT-CFP-Y>F-PIPRSP, or LAT-CFP-Y>F-AIARSA cells were loaded with the calcium indicator dye Indo-I (1 μM) at 37 °C for 30 min. After loading cells with Indo-I, cells were rinsed twice, and resuspended in PBS buffer supplemented with 2% FBS. Cells were incubated with 1:100 dilution of OVA or T4-loaded biotinylated pMHC monomers (NIH Tetramer Core Facility, Atlanta, GA) at 37 °C for 10 min in the prewarmed water bath. Calcium responses were measured and recorded on a LSRFortessa (BD Biosciences). In brief, calcium traces were first recorded for 30 s to obtain the basal calcium levels. Streptavidin was added to the cells at the 30th second to initiate the TCR stimulation. Calcium traces were recorded for a total of 5 min. Data was analyzed with FlowJo software (FlowJo; version 9.9.6).
Imaging Analyses.
More methodologic details can be found in O’Donoghue et al. (51) and Lin et al. (52). The MCC peptide (ANERADLIAYLKQATK) with a C-term GGSC linker was labeled with a Atto647N fluorophore with maleimide chemistry and purified by HPLC. On day 1, lymph nodes and spleens were harvested from primary AND transgenic mice (The Jackson Laboratory). Tissues were homogenized and stimulated with 2 μM MCC peptide. On day 3, cells were retrovirally transduced using the PLAT-E packaging system (Cell Biolabs), delivering a MSCV-LAT-eGFP construct. Cells were imaged on day 5. On day 4, Atto-MCC was loaded into his-MHC molecules via incubation in a solution of 1% BSA and 1× PBS, and pH was adjusted to 4.5 with citric acid. The following day, loaded MHC was isolated with 10-kDa spin filters (Amicon Ultra). Bilayers with 98% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 2% Ni-DOG bilayers 1,2-dioleoyl-sn-glycero-3-[(N-(5- amino-1-carboxypentyl)iminodiacetic acid)succinyl] (Ni2+-NTA-DOGS) (Avanti Polar Lipids) were prepared from a suspension of single unilamellar vesicles (SUVs) made via tip sonication. Number 1.5 coverslips were etched in piranha (3:1 sulfuric acid:hydrogen peroxide) for 3 min. These coverslips were assembled in Attofluor chambers. SUVs were applied to nitrogen dried coverslips and incubated for 35 min, washed with TBS, incubated with 100 mM NiCl2 for 3 min, and then rinsed with TBS. Bilayers were then blocked with 1× Hepes-buffered saline supplemented with Ca, Mg, and 0.1% BSA. After 40 min of blocking, Atto647-MCC-his-MHC and his-ICAM-1 were incubated with the bilayer at ∼50 pM and 80 nM, respectively. Cells were pipetted into the chamber and imaged for 3 min after landing. Detection and tracking were done with TrackMate (75), and analysis was performed with custom Python code.
Supplementary Material
Acknowledgments
We thank the NIH Tetramer Core Facility for providing the OVA and APL peptide-loaded H-2Kb monomers. The work was supported by the Jane Coffin Childs Fund 61-1560 (to W.-L.L.), the HHMI (A.W.) and NIH–National Institute of Allergy and Infectious Diseases Grant P01 AI091580-06 (to J.T.G., A.W., and A.K.C.) and 1R37AI114575 (to A.W.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008303117/-/DCSupplemental.
Data Availability.
Simulation code to calculate channel capacity and error rates of the T cell signaling network is found in the publicly accessible GitHub repository, https://github.com/rganti/Channel_Capacity_T_Cell. All data to understand and access the conclusions of this study are available in the main text, SI Appendix, and the Channel_Capacity_T_Cell GitHub repository.
References
- 1.François P., Altan-Bonnet G., The case for absolute ligand discrimination: Modeling information processing and decision by immune T cells. J. Stat. Phys. 162, 1130–1152 (2016). [Google Scholar]
- 2.Sykulev Y., Joo M., Vturina I., Tsomides T. J., Eisen H. N., Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity 4, 565–571 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty A. K., Weiss A., Insights into the initiation of TCR signaling. Nat. Immunol. 15, 798–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J. et al., A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4+ T cells. Immunity 39, 846–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irvine D. J., Purbhoo M. A., Krogsgaard M., Davis M. M., Direct observation of ligand recognition by T cells. Nature 419, 845–849 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Cole D. K. et al., Human TCR-binding affinity is governed by MHC class restriction. J. Immunol. 178, 5727–5734 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Donermeyer D. L., Weber K. S., Kranz D. M., Allen P. M., The study of high-affinity TCRs reveals duality in T cell recognition of antigen: Specificity and degeneracy. J. Immunol. 177, 6911–6919 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Das J. et al., Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell 136, 337–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stepanek O. et al., Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell 159, 333–345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Germain R. N., Stefanová I., The dynamics of T cell receptor signaling: Complex orchestration and the key roles of tempo and cooperation. Annu. Rev. Immunol. 17, 467–522 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Davis S. J., van der Merwe P. A., The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 7, 803–809 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Germain R. N., Computational analysis of T cell receptor signaling and ligand discrimination—past, present, and future. FEBS Lett. 584, 4814–4822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germain R. N., The T cell receptor for antigen: Signaling and ligand discrimination. J. Biol. Chem. 276, 35223–35226 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Dushek O., Das R., Coombs D., A role for rebinding in rapid and reliable T cell responses to antigen. PLoS Comput. Biol. 5, e1000578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombs D., Dushek O., van der Merwe P. A., “A review of mathematical models for T cell receptor triggering and antigen discrimination” in Mathematical Models and Immune Cell Biology, Molina-París C., Lythe G., Eds. (Springer, 2011), pp. 25–45. [Google Scholar]
- 16.Hopfield J. J., Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U.S.A. 71, 4135–4139 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ninio J., Kinetic amplification of enzyme discrimination. Biochimie 57, 587–595 (1975). [DOI] [PubMed] [Google Scholar]
- 18.McKeithan T. W., Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl. Acad. Sci. U.S.A. 92, 5042–5046 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B., Chen W., Evavold B. D., Zhu C., Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huppa J. B. et al., TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963–967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J. et al., The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 464, 932–936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tischer D. K., Weiner O. D., Light-based tuning of ligand half-life supports kinetic proofreading model of T cell signaling. eLife 8, e42498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yousefi O. S. et al., Optogenetic control shows that kinetic proofreading regulates the activity of the T cell receptor. eLife 8, e42475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss A., Imboden J., Shoback D., Stobo J., Role of T3 surface molecules in human T-cell activation: T3-dependent activation results in an increase in cytoplasmic free calcium. Proc. Natl. Acad. Sci. U.S.A. 81, 4169–4173 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imboden J. B., Weiss A., Stobo J. D., The antigen receptor on a human T cell line initiates activation by increasing cytoplasmic free calcium. J. Immunol. 134, 663–665 (1985). [PubMed] [Google Scholar]
- 26.Mustelin T., Coggeshall K. M., Isakov N., Altman A., T cell antigen receptor-mediated activation of phospholipase C requires tyrosine phosphorylation. Science 247, 1584–1587 (1990). [DOI] [PubMed] [Google Scholar]
- 27.Roose J. P., Mollenauer M., Gupta V. A., Stone J., Weiss A., A diacylglycerol-protein kinase C-RasGRP1 pathway directs Ras activation upon antigen receptor stimulation of T cells. Mol. Cell. Biol. 25, 4426–4441 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo W. L. et al., Slow phosphorylation of a tyrosine residue in LAT optimizes T cell ligand discrimination. Nat. Immunol. 20, 1481–1493 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keane J. F., Atlas L., . “Molecular signal processing and detection in T cells” in 2001 IEEE International Conference on Acoustics Speech, Signal Processing Proceedings, (IEEE, 2001), Vol. 2, pp. 1257–1260. [Google Scholar]
- 30.Keane J. F., Atlas L., . “Signaling efficiency of kinetic proofreading in T-cells” in Genomic signal process. Statistics 2002. GENSiPS 2002. IEEE International Workshop, (IEEE, 2002), Vol. 2, pp. 1597–1600. [Google Scholar]
- 31.Lalanne J. B., François P., Principles of adaptive sorting revealed by in silico evolution. Phys. Rev. Lett. 110, 218102 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Mora T., Physical limit to concentration sensing amid spurious ligands. Phys. Rev. Lett. 115, 038102 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Cover T. M., Thomas J. A., Elements of Information Theory, (Wiley, 2012). [Google Scholar]
- 34.Altan-Bonnet G., Germain R. N., Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 3, e356 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefanová I. et al., TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat. Immunol. 4, 248–254 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Lipniacki T., Hat B., Faeder J. R., Hlavacek W. S., Stochastic effects and bistability in T cell receptor signaling. J. Theor. Biol. 254, 110–122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lever M. et al., Architecture of a minimal signaling pathway explains the T-cell response to a 1 million-fold variation in antigen affinity and dose. Proc. Natl. Acad. Sci. U.S.A. 113, E6630–E6638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bel G., Munsky B., Nemenman I., The simplicity of completion time distributions for common complex biochemical processes. Phys. Biol. 7, 016003 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Murugan A., Huse D. A., Leibler S., Speed, dissipation, and error in kinetic proofreading. Proc. Natl. Acad. Sci. U.S.A. 109, 12034–12039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nika K. et al., Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity 32, 766–777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatada M. H. et al., Molecular basis for interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature 377, 32–38 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Thill P. A., Weiss A., Chakraborty A. K., Phosphorylation of a tyrosine residue on Zap70 by Lck and its subsequent binding via an SH2 domain may be a key gatekeeper of T cell receptor signaling in vivo. Mol. Cell. Biol. 36, 2396–2402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo W. L. et al., Lck promotes Zap70-dependent LAT phosphorylation by bridging Zap70 to LAT. Nat. Immunol. 19, 733–741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez C. F., Muhlich J. L., Bachman J. A., Sorger P. K., Programming biological models in Python using PySB. Mol. Syst. Biol. 9, 646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown P. N., Byrne G. D., Hindmarsh A. C., VODE: A variable-coefficient ODE solver. SIAM J. Sci. Stat. Comput. 10, 1038–1051 (1989). [Google Scholar]
- 46.Houtman J. C. D., Houghtling R. A., Barda-Saad M., Toda Y., Samelson L. E., Early phosphorylation kinetics of proteins involved in proximal TCR-mediated signaling pathways. J. Immunol. 175, 2449–2458 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J., Weiss A., Identification of the minimal tyrosine residues required for linker for activation of T cell function. J. Biol. Chem. 276, 29588–29595 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Shah N. H. et al., An electrostatic selection mechanism controls sequential kinase signaling downstream of the T cell receptor. eLife 5, e20105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katz Z. B., Novotná L., Blount A., Lillemeier B. F., A cycle of Zap70 kinase activation and release from the TCR amplifies and disperses antigenic stimuli. Nat. Immunol. 18, 86–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi J., Balagopalan L., Nguyen T., McIntire K. M., Samelson L. E., TCR microclusters form spatially segregated domains and sequentially assemble in calcium-dependent kinetic steps. Nat. Commun. 10, 277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Donoghue G. P., Pielak R. M., Smoligovets A. A., Lin J. J., Groves J. T., Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife 2, e00778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J. J. Y. et al., Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci. Signal. 12, eaat8715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang W. Y. C. et al., Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc. Natl. Acad. Sci. U.S.A. 113, 8218–8223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang W. Y. C. et al., A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniels M. A. et al., Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Hartgroves L. C. et al., Synergistic assembly of linker for activation of T cells signaling protein complexes in T cell plasma membrane domains. J. Biol. Chem. 278, 20389–20394 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Genot E., Cantrell D. A., Ras regulation and function in lymphocytes. Curr. Opin. Immunol. 12, 289–294 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Margarit S. M. et al., Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Gureasko J. et al., Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 15, 452–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iversen L. et al., Molecular kinetics. Ras activation by SOS: Allosteric regulation by altered fluctuation dynamics. Science 345, 50–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Courtney A. H. et al., A phosphosite within the SH2 domain of Lck regulates its activation by CD45. Mol. Cell 67, 498–511.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodfellow H. S. et al., The catalytic activity of the kinase ZAP-70 mediates basal signaling and negative feedback of the T cell receptor pathway. Sci. Signal. 8, ra49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paster W. et al., A THEMIS:SHP1 complex promotes T-cell survival. EMBO J. 34, 393–409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Courtney A. H., Lo W. L., Weiss A., TCR signaling: Mechanisms of initiation and propagation. Trends Biochem. Sci. 43, 108–123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parrondo J. M. R., Horowitz J. M., Sagawa T., Thermodynamics of information. Nat. Phys. 11, 131–139 (2015). [Google Scholar]
- 66.Hogenboom D. L., Maxwell’s demon: Entropy, information, computing. Am. J. Phys. 60, 282–283 (1992). [Google Scholar]
- 67.Williams B. L. et al., Phosphorylation of Tyr319 in ZAP-70 is required for T-cell antigen receptor-dependent phospholipase C-gamma1 and Ras activation. EMBO J. 18, 1832–1844 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pelosi M. et al., Tyrosine 319 in the interdomain B of ZAP-70 is a binding site for the Src homology 2 domain of Lck. J. Biol. Chem. 274, 14229–14237 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Wang H. et al., ZAP-70: An essential kinase in T-cell signaling. Cold Spring Harb. Perspect. Biol. 2, a002279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Courtney A. H. et al., CD45 functions as a signaling gatekeeper in T cells. Sci. Signal. 12, eaaw8151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balagopalan L., Kortum R. L., Coussens N. P., Barr V. A., Samelson L. E., The linker for activation of T cells (LAT) signaling hub: From signaling complexes to microclusters. J. Biol. Chem. 290, 26422–26429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nag A., Monine M. I., Faeder J. R., Goldstein B., Aggregation of membrane proteins by cytosolic cross-linkers: Theory and simulation of the LAT-Grb2-SOS1 system. Biophys. J. 96, 2604–2623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nag A., Monine M., Perelson A. S., Goldstein B., Modeling and simulation of aggregation of membrane protein LAT with molecular variability in the number of binding sites for cytosolic Grb2-SOS1-Grb2. PLoS One 7, e28758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Houtman J. C. D. et al., Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat. Struct. Mol. Biol. 13, 798–805 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Tinevez J. Y. et al., TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Simulation code to calculate channel capacity and error rates of the T cell signaling network is found in the publicly accessible GitHub repository, https://github.com/rganti/Channel_Capacity_T_Cell. All data to understand and access the conclusions of this study are available in the main text, SI Appendix, and the Channel_Capacity_T_Cell GitHub repository.