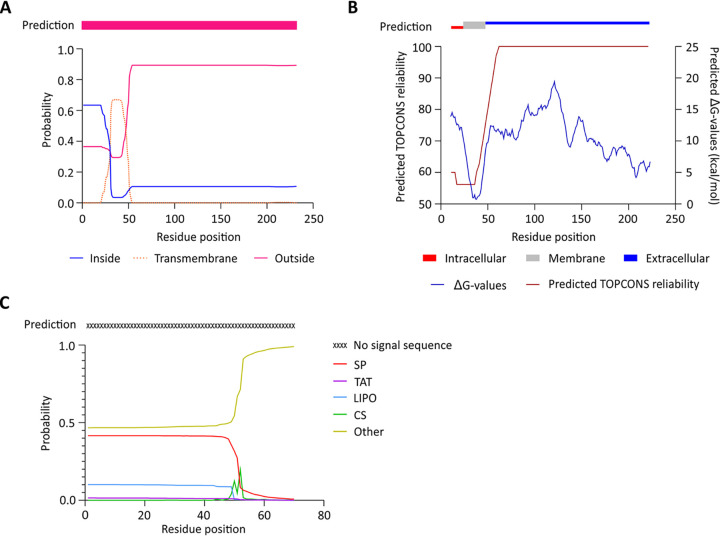
FIG 1.
Prediction of a transmembrane helix in TcdC. (A) Output from the prediction by TMHMM 2.0 software (24) through the 1-best algorithm (pink bar) and probability plot for the area inside the cell, the transmembrane region, and the area outside the cell. (B) Prediction by TOPCONS software (25), with consensus in residues 1 to 26 inside the cell, a transmembrane helix (residues 26 to 46), and residues 47 to 232 on the outside of the cell. The TOPCONS reliability score and predicted ΔG values for each residue are shown. (C) Output from the SignalP 5.0 (26) web server for the TcdC amino acid sequence. The probabilities of signal peptide presence from the systems Sec (SP), TAT, and lipoprotein (LIPO) are shown. The predicted cleavage site score (CS) and no signal sequence probability (Other) are depicted.