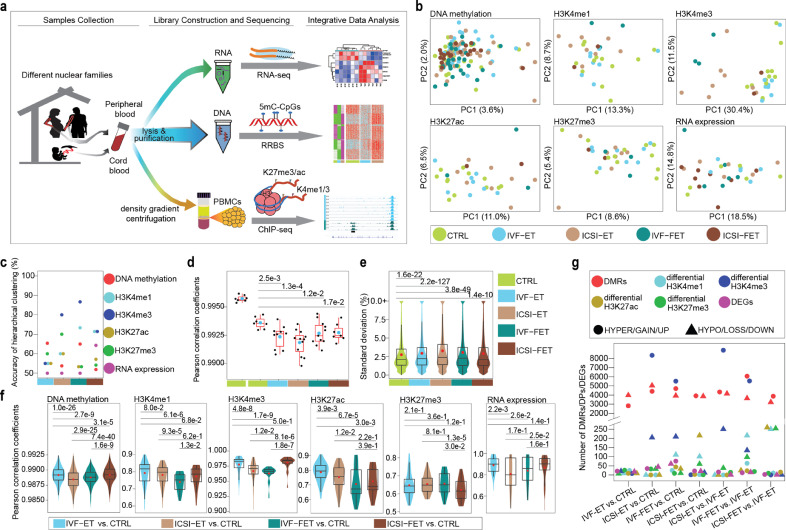
Fig. 1.
Epigenome profiling in ART-conceived neonates. (a) Graphical overview of the study design. (b) Principal component analysis (PCA) for all the five groups (CTRL, IVF-ET, ICSI-ET, IVF-FET, and ICSI-FET) of neonatal samples by using each layer of the reference epigenome and transcriptome. For DNA methylation (sample size: n = 137), only the 100-bp tiles covered all neonatal samples were used; ChIP-seq peaks of all neonatal samples (sample size: n = 33) for each histone modification were pooled together, then the overlapping peaks were merged; genes with TPM=0 in all neonatal samples(sample size: n = 32) are removed. Finally, the number of genomic regions for each epigenomic layer to generate the PCAs: DNA methylation (467,097 100-bp tiles), H3K4me1 (195,570 peaks), H3K4me3 (86,157 peaks), H3K27ac (324,900 peaks), H3K27me3 (257,175 peaks), and RNA expression (26,326 genes). (c) The accuracy of hierarchical clustering for IVF-ET versus CTRL (cyan), ICSI-ET versus CTRL (sienna), IVF-FET versus CTRL (dark cyan), and ICSI-FET versus CTRL (dark sienna) neonatal samples. Samples and genomic regions used were the same as in (b). (d) Box plots for the distribution of the within-twin-pair Pearson correlation coefficients for genome-wide DNA methylation in each group as in (b), respectively. Twin pairs in CTRL group were classified into two subgroups, monozygotic and dizygotic twins. Each black dot represents the Pearson correlation coefficient and the cyan dots are the arithmetic means. The p-value between two groups was determined by unpaired and two-tailed t-test. (e) Violin-box plots showed the distribution of standard deviations of genome-wide DNA methylation for neonatal samples in each group mentioned in (b), respectively. The p-value between two groups was determined by Wilcoxon rank-sum test. (f) For each layer of the reference epigenome and transcriptome, violin-box plots showed the distribution of Pearson correlation coefficients between CTRL and one of the four ART groups. The red dots are the arithmetic means. The p-value between two groups was determined by Wilcoxon rank-sum test. (g) The number of DMRs, DPs of four histone modifications, and DEGs for neonatal samples were shown for seven comparisons: IVF-ET versus CTRL, ICSI-ET versus CTRL, IVF-FET versus CTRL, ICSI-FET versus CTRL, ICSI-ET versus IVF-ET, IVF-FET versus IVF-ET, and ICSI-FET versus IVF-ET.